Introduction
Diabetes mellitus is a chronic metabolic disease that is associated with elevated blood sugar concentrations due to impaired insulin production or effectiveness., It is classified into two types. These two forms of the disease are Type 1 diabetes mellitus (T1DM) and Type 2 diabetes mellitus (T2DM)., Gestational diabetes and other specific types are also known, though they are less frequent. T1DM is an autoimmune disease in which the body’s immune system attacks and destroys insulin-secreting beta cells in the pancreas, resulting in a lack of insulin. It is a condition most commonly diagnosed in children and adolescents, but it can affect anyone at any age. Patients need to take insulin for the rest of their lives in order to survive. T2DM is a condition characterized by insulin resistance and insufficient insulin secretion. Obesity, physical inactivity, and an unhealthy diet closely correlate with T2DM, affecting more adults., Genetic factors also influence it., While insulin injections are the only way to regulate blood sugar in T1DM, changes in diet, exercise, and oral agents can treat T2DM, which is more frequent., Unmanaged diabetes can lead to complications such as microvascular and macrovascular diseases, increased susceptibility to infections, diabetic foot ulcers, and skin conditions.,
Diabetes mellitus is a widespread health issue that affects millions of people globally, with 537 million adults affected globally. If the current trends persist, this figure will reach 643 million by 2030 and 783 million by 2045, according to current estimates. T2DM affects 90–95% of the diabetes population, while T1DM is also on the rise, especially in the developed world. Diabetes is one of the most pressing health issues of the present time, which has major implications for healthcare systems and demands actions regarding prevention, early identification, and management. A timely and correct diagnosis is critical to managing and preventing related complications. The current diagnosis of diabetes is primarily based on blood glucose testing and glycated hemoglobin (HbA1c) levels. However, these approaches’ limitations include reduced sensitivity and specificity, especially in the initial stages of the disease.
RNA-based diagnostics present a revolutionary approach to the diagnosis of diseases and help in the identification of disease developments as well as the development of individualised treatments. These diagnostics are based on the RNA patterns of gene expression, which can better mirror the actual physiological conditions and disease conditions than conventional biomarkers. The specificity and opportunities for individualization that RNA-based diagnostics present could transform diabetes management, resulting in improved outcomes and lower costs.
Despite the great potential of RNA-based diagnostics in the diagnostics field, unlocking their full potential requires overcoming several issues. These include assessing the specificity of RNA biomarkers, developing accurate and reproducible diagnostic approaches, and addressing practical scenarios such as affordability, availability, and compatibility with routine clinic work. It is thus important and appropriate to present a comprehensive review of RNA-based diagnostics in diabetes care. This review will thus aim at offering a detailed review of the current literature, the identified gaps, and the potential future research directions. This way, it seeks to close the gap between new molecular diagnostic developments and their implementation in diabetes care, with the overall goal of enhancing patient care.
Methodology
Literature search
We conducted an extensive search in various databases, such as PubMed, Google Scholar, Web of Science, and Scopus. We used the following search terms: RNA biomarkers, diabetes diagnosis, microRNAs, long non-coding RNAs, messenger RNAs, diabetes monitoring, personalised medicine, diabetes management, molecular diagnostics, and clinical application. We limited the search to English articles published between 2014 and 2024.
Inclusion and exclusion criteria
Inclusion criteria:
- Articles from peer-reviewed journals, well-controlled clinical trials, and high-level review articles.
- Research that addresses the participation of RNA molecules in the detection, assessment, and treatment of diabetes mellitus.
- Scientific papers describing the use of RNA-based diagnostics in the treatment of diabetes and the advantages and disadvantages of their application.
Exclusion criteria:
- Articles, editorials, and opinion columns that are not published in scholarly journals.
- Other research that is not specific to the development of RNA-based diagnostics for diabetes.
- Articles that have been published prior to the year 2015
Data extraction
Relevant data were extracted from the selected articles, including:
- The specific form of RNA molecules that are discussed (for example, microRNA, long non-coding RNA, or mRNA).
- Techniques employed for the identification and estimation of RNA.
- The role of RNA biomarkers in diagnostics and prediction of the disease course.
- Clinical implications and uses in the management of diabetes mellitus.
- Challenges and shortcomings observed in the studies.
Data analysis and synthesis
We synthesized the obtained data to identify the common outcomes, directions, and shortcomings of the current investigations. We grouped the articles based on the type of RNA molecule involved, its role in diabetes management, and the methods used. We compiled the data to provide a comprehensive understanding of the current state of RNA for diagnostic purposes in diabetes care. This approach allows for a comprehensive and structured analysis of the literature on RNA-based diagnostics in diabetes care, as well as offering important suggestions for further research and practical application.
Types of RNA molecules in diabetes diagnostics
MicroRNAs (miRNAs)
MicroRNAs, or miRNAs, are short, non-coding RNAs that usually have 18 to 25 nucleotides., They control gene expression after transcription by attaching to specific sequences in target mRNAs and stopping translation or destroying the mRNA. Many research studies have described certain miRNA signatures that are characteristic of T1DM and T2DM. For instance, miR-375 and miR-126., T1DM and T2DM dysregulate the expression level of miR-375, which plays a role in pancreatic beta-cell development and insulin secretion. miR-126 is involved in endothelial function and insulin action. Decreased levels are associated with T2DM and its vascular complications. Molecular markers, such as circulating miRNAs in the blood and other biofluids, can be useful in the diagnosis of diabetes and the assessment of its course. According to Kamalden et al., miR-15 has a critical role in insulin release in pancreatic β-cells and plays a role in retinal damage during the development of type 2 diabetes. According to Jimenez-Lucena et al. study, HbA1c and circulating miRNA levels may function as predictive biomarkers for the development of T2DM in individuals with coronary heart disease.
Long non-coding RNAs (lncRNAs)
Long non-coding RNAs (lncRNAs) are defined as RNA molecules containing more than 200 nucleotides and not coding for proteins. They are also involved in controlling gene expression at different levels, such as the modification of chromatin, transcription, and post-transcription, though not all lncRNA have a functional purpose. lncRNAs have been found to be dysregulated in Type 2 Diabetes. For instance, lncRNA H19 is connected with insulin resistance and diabetic complications, including retinopathy and nephropathy; thus, it can be useful in detecting and assessing complications. Fan et al. Found that by suppressing the expression of the vitamin D receptor, a negative feedback loop involving H19/miR-675/EGR1 contributes to diabetic nephropathy. LncRNA metastasis-associated lung carcinoma transcript 1 (MALAT1) is implicated in endothelial cell dysfunction and inflammation in diabetes. Lorenzen and Thum suggest that inhibiting MALAT1 expression can maintain normal inflammatory factors like IL6 and TNF-a, thereby reducing diabetes-related complications. LncRNA MALAT1 activates the p38 mitogen-activated protein kinase signalling pathway in diabetic cataract, hence promoting oxidative stress and death of human lens epithelial cells. Therefore, lncRNAs can help identify the risk and timing of the development of diabetic complications. LncRNAs also change the expression related with pathways of oxidative stress, inflammation and fibrosis in diabetic complications. It is proposed that they have altered levels in diabetic patients, and as such, are potential early markers for the progression of diabetes., It is also proposed that all lncRNAs possess tissue specificity, implying that they may be specific to particular organs. They may also communicate with other molecules, for example microRNAs or proteins and can regulate the action of such molecules. LncRNAs are implicated in epigenetic regulatory mechanisms such as DNA methylation and histone modification. The said observations result in better identification of developmental complications and corresponding treatments that may minimize or halt complications at their earliest stages.Thus, regulating lncRNA levels may represent a novel strategy for minimizing or managing complications associated with Diabetes
Circular RNAs (circRNAs)
Circular RNAs (circRNAs) are a class of noncoding RNAs that form a covalent closed loop structure, which is more stable than linear RNAs. It can serve as a miRNA sponge, regulate transcription, and bind RNA-binding proteins. CircRNAs are involved in gene regulation and are known to play a role in diabetes development., They are stable and easily available in body fluids, and thus, excellent candidates for non-invasive diabetes diagnosis and management. The insulin gene’s circRNA, ci-Ins2/ci-INS, was discovered by Stoll and colleagues. It interacts with the 43-kDa TAR DNA-binding protein to control insulin release. The islets of rats and individuals with type 2 diabetes have decreased levels of ci-Ins2/ci-INS.
CircHIPK3 plays a role in insulin secretion and beta-cell proliferation, while CircCAMSAP1 has implications for insulin resistance and glucose homeostasis. Shan et al. reported that retinal vascular dysfunction in diabetes mellitus is mediated by circHIPK3. CircHIPK3 regulates pancreatic β-cell insulin production and insulin mRNA levels, according to research by Stoll and colleagues. Knowledge of circRNA functions can be useful in elucidating diabetes pathogenesis.
Messenger RNAs (mRNAs)
mRNAs are RNA molecules that transport genetic information from DNA to the ribosome, facilitating the synthesis of proteins. Changes in the levels of mRNA are useful in determining the functional state of cells, especially pancreatic beta cells, in the case of diabetes. Most importantly, changes in glucagon mRNA levels can represent alpha-cell function and glucose balance. The development of glucagon physiology indicates that it is involved in multiple physiological mechanisms that control energy balance and glucose homeostasis. Gene expression studies on the mRNA populations in samples taken from diabetic patients can reveal gene expression alterations involved in the development of diabetes and its progression. Therefore, the measurement of mRNA levels can help evaluate the effectiveness of diabetes monitoring and therapies.
Exosomal RNAs in diabetes diagnostics
Exosomes are small vesicles with a size ranging from 30 to 150 nm that are secreted by cells and contain molecules such as RNA, proteins, and lipids. The RNA content of exosomes reflects the cells in which they are located, so studying them can aid diagnosis. Thus, exosomal RNAs derived from blood can be useful for diagnosing diabetes and its development and can be considered as potential biomarkers. The functions of beta cells, insulin resistance, and diabetic complications are associated with some of the miRNAs in the exosomes. In diabetes, exosomal lncRNAs and circRNAs are involved in the evaluation of cellular stress and metabolic alterations. According to Sun et al.’s research, exosomes produced from human mesenchymal stem cells reverse peripheral insulin resistance and reduce β-cell death to mitigate type 2 diabetes mellitus. Similarly, Sun et al., observed that in the islets of streptozotocin-induced diabetic mice, exosomes from β-cells reduced hyperglycemia and increased angiogenesis. Thus, exosomal RNAs derived from blood or urine samples of diabetic patients may be utilised as diagnostic and prognostic markers of the disease. This can be helpful in tracking the disease’s progression and treatment success.
Single-cell RNA sequencing (scRNA-seq) in diabetes research
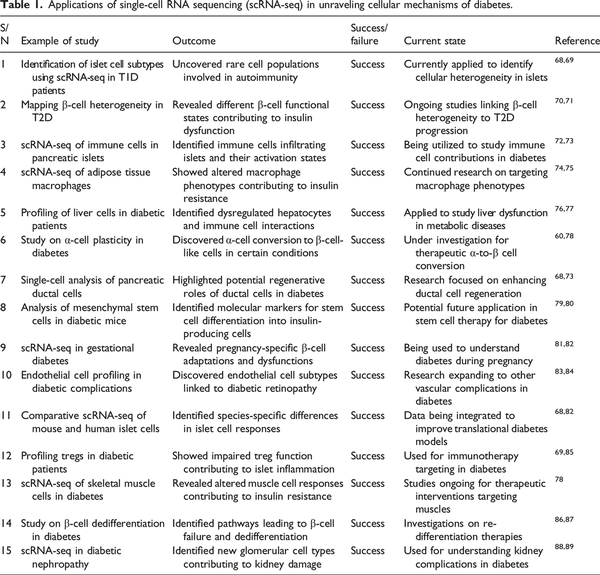
The Single-cell RNA sequencing (scRNA-seq) process includes capturing single cells, converting the RNA to complementary DNA (cDNA), and then sequencing to obtain gene expression patterns in cells. Some of the techniques include droplet-based methods such as 10x Genomics, microfluidic-based systems such as Fluidigm C1, and plate-based techniques such as SMART-seq. scRNA-seq helps in the examination of gene expression at the single-cell level and therefore captures cellular diversity. In diabetes research, scRNA-seq has been applied to identify pancreatic islet cells, their function in diabetic conditions, and the subpopulation of beta cells. This technology helps in discovering new biomarkers and the pathophysiology of beta-cell failure. scRNA-seq is a technique that has recently transformed the study of cellular heterogeneity and gene expression in individual cells. Conventional RNA sequencing gives an overall gene expression pattern for a population of cells, which may hide important variations between cells. This disadvantage is solved by single-cell RNA sequencing, which allows analysing the gene expression of single cells and thus identifying cellular heterogeneity and certain transcriptional states. Specifically, scRNA-seq has been applied to study pancreatic islet cell heterogeneity, the function of beta-cells in diabetes, the discovery of new biomarkers, and the development of therapeutic interventions. It is useful in establishing early markers of beta-cell stress and dysfunction, disease progression, and complications in the kidney and retinal cells.Table 1 below indicate applications of single-cell rna sequencing (scRNA-seq) in unraveling cellular mechanisms of diabetes.
The limitations or challenges of using single cell RNA sequencing
Single-cell RNA sequencing (scRNA-seq) is a powerful tool for studying gene expression at the single-cell level, but it comes with several limitations and challenges,:
High cost and technical complexity
scRNA-seq is costly mainly because it needs expensive instruments, chemicals, and computation facilities. This includes steps that include cell isolation, RNA capture and sequencing which are time consuming, technical and needs optimization.
Low RNA quantity
In individual cells, RNA is in relatively limited availability; hence, it can be difficult to acquire well-defined responses. Therefore, new amplification steps are needed, but these are often accompanied by biases and noise.
Dropout events
Dropout is defined as the inability of an experimental technique to identify RNA transcripts that are actually present in a cell at a given time. This is a well-known problem for scRNA-seq, where lowly expressed genes are often not detected but identification of their absence may provide misleading information.
Batch effects
Inter-sample or inter-run variation can generate batch effects disrupting the differentiation of biological contrasts. This, in turn, entails appropriate design of experiments and normalisations of acquired data properly.
Cell heterogeneity
Analysed samples include scRNA-seq which is used to produce big data from different cell types yet the data is challenging to analyse due to complexity in biological samples. It always poses a challenge to distinguish between biological variation and noise sources.
Data processing and interpretation
scRNA-seq data analysis and subsequent interpretation processes involve some complex computations which can only be handled by experts in computational biology. The data generated can sometimes be huge and requires strong computational processing, normalization and visualization.
Cell isolation and viability
Single cell sorting may adversely affect the cells or change their transcriptional profiles which may in turn affect the outcome. Also, some cells are hypothesized to be more sensitive or harder to capture and hence impacts the nature of cells that are measured.
Limited spatial information
Current scRNA-seq approaches do not incorporate any information about the spatial position of the cells in the tissue. This has hampered analysis of cell – cell interactions and tissues organization although new technologies such as spatial transcriptomics are gradually filling this gap.
Technical artifacts
Sequencing and library preparation processes themselves can generate artifacts such as overexpression of specific transcripts which makes the results biases.
RNA-based monitoring of glycemic control
Glycemic control is an essential component of diabetic mellitus care to prevent complications using blood glucose level regulation. Widely used tests such as HbA1c, fasting blood glucose, and continuous glucose monitoring (CGM) have their own limitations. Other techniques that are based on RNA, for instance, miRNA and mRNA, can enhance the conventional approaches by providing real-time information on glucose and insulin functions. They are able to assess fluctuations in glucose concentrations and any abnormalities in glucose regulation. During hyperglycemia, characterized by high intracellular glucose levels, the expression of certain RNA molecules rises to promote insulin release or glucose absorption. Conversely, RNA-mediated responses during hypoglycemia may stimulate glucose synthesis or inhibit insulin secretion.– They are associated with pancreatic β-cell development and insulin secretion function, and the levels of miRNAs have been shown to be associated with the function of β-cells and glycemic control.
LncRNAs are implicated in insulin resistance and diabetic complications, and the levels of their expression correlate with glucose metabolism and insulin sensitivity. Therefore, lncRNAs can contribute to the regulation of blood glucose fluctuations and long-term glycemic control, and they may also have the potential to improve insulin sensitivity and glucose management. circRNAs can act as miRNA sponges, regulate transcription, and perform other processes with RNA-binding proteins. As a result, they are potential biomarkers for the long-term and sensitive evaluation of glycemic control, and they describe the ways and means of glucose metabolism regulation.
circRNAs are involved in glycemic regulation since they act as miRNA sponges thus modulating the activity of specific miRNAs that appear to impact on genes. Specifically, these circRNAs alter the pathways in insulin secretion, insulin sensitivity, glucose transport and utilization. These also modulate β-cell function a process that plays a very sensitive role in the secretion of insulin. It has been identified that CirRNAs have the potential to be biomarkers of glycemic control, because they are stable molecules present in body fluids, such as blood, and their detection does not require invasive procedures. They can also be used to assess short-term glycemic fluctuations or long-term glycemic control for providing prognostication about complications arising out of diabetes. Some circRNAs are involved in the development of insulin resistance; therefore, enhancing or suppressing the expression of the circRNAs can enhance insulin sensitivity of the cells. The potential of circRNA-based treatments could be examined; one idea is to create synthetic circRNAs to replace the disrupted circRNAs or use small molecules to block the binding of circRNAs to their targets.
RNA-based monitoring of glycemic control has drawbacks, such as a lag in reflecting glucose levels, the impact of haemoglobin variations, and the absence of real-time data. HbA1c, a form of glycated haemoglobin, indicates blood glucose regulation during the preceding 2-3 months, although it is not effective for monitoring daily variations. Fasting Blood Glucose (FBG) provides a single measurement but fails to reflect glucose regulation throughout the course of the day. Transient variables such as dietary modifications, physical activity, stress, sickness, and sleep may skew FBG values. FBG is reactive to instances of hypoglycemia outside the fasting interval. Continuous Glucose Monitoring (CGM) necessitates sensor calibration, introducing inaccuracies, and exhibits a lag time of 10–15 min, particularly during glycaemic swings., Implantable continuous glucose monitoring (CGM) is less expensive than continuous subcutaneous glucose monitoring (SCGM) but remains unaffordable for some patients, particularly in poor nations. These constraints underscore the need for a focused strategy in analysing glucose monitoring data.
RNA-based diagnostics for diabetic foot ulcers
A diabetic foot ulcer (DFU) is a persistent wound that does not heal, affects the quality of life of diabetic patients, and can become complicated and result in amputations. Current diagnostic approaches, such as clinical assessment and imaging, have several limitations and fail to show the necessary level of sensitivity and specificity for the identification and tracking of DFUs. RNA-based diagnostics are a promising concept as they rely on the stability and specificity of RNA molecules to understand the molecular processes of DFU development and progression.
New RNA-based diagnostic approaches, including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA), have shown potential for enhancing the diagnosis and prognosis of DFUs. MiRNAs control gene expression at the post-transcriptional level and are found to be downregulated in DFUs, thus suggesting poor angiogenesis and wound healing. It has been reported that altered levels are found in DFUs and are related to chronic inflammation and poor healing of DFUs. DFUs are associated with low levels of miR-15b, miR-21, and miR-146a, all of which are implicated in angiogenesis, inflammation, and cell proliferation. These downregulations can cause effects such as inhibition of angiogenesis, inflammation, and delayed healing. Circulating miRNAs can be used as biomarkers for the early diagnosis and assessment of DFUs, and certain miRNA signatures can be used to predict the chances of healing and the chances of complications. It has been proven that lncRNAs can participate in gene expression in different ways, and it has been postulated that controlling dysregulated lncRNAs could be beneficial in the enhancement of wound healing. Excessive MALAT1 and H19 expression linked to chronic wound pathology, cell proliferation, and migration is reported to be connected with the formation of DFUs. These lncRNAs may act as diagnostic markers and therapeutic targets, thus helping to heal wounds and discover new treatments. The altered levels are associated with impaired wound healing in the DFUs, while the dysregulated expression is associated with chronic inflammation and a slow healing rate. CircRNA-HIPK3 and CircRNA-FOXO3 regulate cell proliferation and migration in DFUs and, thus, the wound healing process. If left unchecked, it leads to inflammation and poor healing of the tissues. CircRNAs have the potential to be used as diagnostic markers, help in monitoring the progression of DFUs, and may provide some insights into the molecular mechanisms that underlie DFU development. Understanding the functions of circRNA can contribute to the understanding of the molecular mechanism of DFU development, and treat.
RNA interference (RNAi) for treatment monitoring
RNA interference (RNAi) is a gene regulation mechanism in which RNA molecules inhibit gene expression, usually by mediating the degradation of target mRNA., This mechanism has been understood to be very efficient in the management and even prevention of various diseases, such as diabetes mellitus. Glycemic control is vital in the management of diabetes in order to avoid other complications that include cardiovascular diseases, neuropathies, nephropathies, and retinopathies. Through the manipulation of genes related to insulin production, insulin sensitivity, and inflammation, RNAi can modulate the manifestation of diabetes.
RNAi has several steps, including dicer processing, loading of the RNA-induced silencing complex, and target recognition and cleavage. The guide strand directs the RNA-induced silencing complex to bind to the complementary mRNA, which stops translation of the mRNA. RNAi can be used to treat diabetes through gene knockout. SiRNA regulates the insulin gene, influencing the enzymes involved in pro-insulin synthesis. This may help raise the secretion of insulin in pancreatic beta cells. SiRNA, which controls the PI3K/Akt pathway, can upregulate the insulin receptor gene, thereby increasing insulin signalling and glucose transport. This can be done either through the silencing of the genes or by altering the levels of the miRNAs. Additionally, siRNA targeting anti-inflammatory cytokines could potentially alleviate the inflammation associated with diabetic conditions. The above can be achieved by siRNA gene knockdown of the pro-inflammatory genes or by altering the level of miRNA in order to regulate inflammation. These approaches are useful in the prevention and control of chronic inflammation, as well as in the enhancement of glycemic control in diabetic patients.
Some of the benefits include high specificity since it can pinpoint diseased genes, flexibility, and being non-invasive. However, the problem of delivering RNAi molecules to certain tissues, such as pancreatic islets, is still a challenge. Stability is an important factor to consider in treatment, and off-target effects are always a big problem that must be considered and analyzed in the best way possible.
Integration with digital health platforms
RNA-based diagnostics is promising if established as very accurate and effective tools for the identification and assessment of diabetes-associated markers. Hence, the combination of specific RNA biomarkers and the real-time data of digital health technologies can leads to more accurate, timely, and effective management and prevention of diabetes. Technologies such as mobile health (m-health), telemedicine, and other digital platforms have revolutionised health care delivery systems as they enhance real-time data capture and telemonitoring of patients. The use of these technologies can enhance the management of individuals with diabetes as it provides an evidence-based view of the patient and diabetes to the patient and healthcare provider.
The diagnostic techniques include next-generation sequencing, quantitative PCR, and microarray analysis for overall characterization, biomarker identification, and validation of specific RNA molecules for screening as well. Some of the trends that are gathering momentum today include the use of smartphones for diagnosis, the storage of patient data in the cloud, and the wearing of biosensors., Smartphone-Integrated Diagnostics will empower users to conduct tests and immediately receive results. Cloud-based data platforms assist in storing and analyzing health data, providing recommendations and forecasts for personalized diabetes treatment. Implantable biosensors check RNA biomarkers in body fluids to monitor the patient’s health status on a regular basis. However, new challenges remain, and technological advancements, data protection concerns, and legal frameworks will shape the continued evolution of the use of such integrated approaches. Digital health technologies and RNA biomarkers can enhance the care and well-being of diabetes patients.
RNA biosensors for point-of-care testing
Point-of-care testing (POCT) which uses RNA biosensors for diabetes management due to their fast, sensitive, and selective nature will improve glucose monitoring in glycemic control. These biosensors can detect specific RNA molecules that are associated with diabetes, thereby enabling real-time intervention in diabetes management. Some common ways to use RNA biosensors are hybridization-based detection, in which the target RNA is linked to a probe sequence that is marked with a fluorescent or electrochemical signal; enzyme-linked detection, in which the target RNA is found with the help of RNA aptamers or other molecules that can start enzymes working; and electrochemical detection, which is used to find out about electrical properties. RNA biosensors are valuable tools in diabetes care because they help evaluate glycemic control, diagnose complications, and monitor patients. The development of the RNA biosensors that detect miR375, miR192 and miR215 are beneficial due to their pancreatic specific expression and thus they play a crucial role in the functional activity of beta cells and glucose regulation. Electrochemical and fluorescent biosensors are the detection techniques. These biomarkers can also be used for the diagnosis of diabetic nephropathy and diabetic retinopathy and for the management of these conditions. The idea of an individual approach to diabetes management according to the RNA profiles makes it possible to fine-tune all of the necessary changes in the therapeutic regimen immediately. Examples include multiplexed biosensors, which detect multiple RNA targets simultaneously, and wearable biosensors, which integrate the biosensors into wearable devices for continuous and real-time monitoring of RNA biomarkers. Some of the advancements in the field of RNA biosensors for diabetes include the use of nanotechnology, nanoparticles, nanowires, nanotubes, microfluidics, point of use, the CRISPR-Cas system, SHERLOCK, and DETECTR. Nonetheless, the current and future trends in research and development suggest their application in clinical practice. The future of RNA biosensors in diabetes POCT is to help achieve better disease control through individualized, quick, and constant monitoring of the patient’s status.
Artificial intelligence and machine learning in developing RNA-based diagnostics
Diabetes mellitus is one of the world’s most pressing and widespread health issues, characterised by elevated blood sugar levels that result from inadequate insulin production, usage, or both. Management of diabetes mellitus entails the monitoring of biomarkers with regards to the glycemic status and the progression of complications. The integration of artificial intelligence (AI) and machine learning (ML) into RNA-based diabetes diagnostics has revolutionised diabetes care. Notably, AI and ML can improve the sensitivity and specificity of RNA-based diagnostic methods, enabling early and personalized treatment of diabetes patients.
Some of the diagnostic methods are next-generation sequencing (NGS), quantitative PCR (qPCR), and microarray analysis. AI and ML are applied in RNA-based diagnostics for biomarker identification, in predictive analytics, in anomaly detection, and in designing individual treatment regimens, as well as in the integration of the RNA data with the clinical data.
In enhancing the discussion on deep learning, autoencoder as well as reinforcement learning applied to RNA-based diagnostics of diabetes, it is pertinent to describe these strategies in detail, predicting the probability of pre-diabetes or the outcome of a transition to diabetes. Deep learning is categorized under machine learning utilizing high-dimensional RNA sequencing data and Artificial Neural Network. This has been applied in identifying microRNA signatures that associate with diabetes thereby increasing the diagnostic yield. Deep learning models demonstrate above 90% of accuracy rates for estimating diabetic condition from RNA expression patterns of large patient data sets.
Autoencoders are the type of generative models that arise in the unsupervised learning context where the input data is used to generate a feature vector of inputs and the reconstruction of the features. In RNA-based diabetes diagnostics, they can minimize the dimensionality of RNA-seq data, selecting only the features that are to be used in disease risk prediction. For instance, they have been used to reduce high dimensional RNA expression data of thousands of genes to a small set of dimensions while still capturing diagnostic presentations of diabetes biomarkers.,
Reinforcement learning, where an algorithm is trained to make decisions and is later rewarded when it produces a correct decision and punished when it produces an incorrect decision is becoming popular in RNA-based diabetes diagnostics. This technique has been employed to improve RNA probe selection in diagnostic tests and refine diagnostic algorithms in use for maximal effectiveness.
Applications of AI can assist in analyzing large data from RNA sequencing to discover new diabetes biomarkers, while ML can select the most suitable set of RNA biomarkers for disease diagnosis and prediction. AI can also be useful in determining the likelihood of disease occurrence, its development, and the outcome of treatment based on RNA biomarkers. This integration improves diagnostic accuracy and patient classification. ML has shown great promise in the early diagnosis of diabetes and its accompanying complications. For example, Shukla achieved an accuracy rate of 82.92% by applying a logistic regression algorithm to predict diabetes risk in Indian individuals using clinical and demographic factors. This suggests that the established model could detect those who are at risk of developing diabetes and possibly even stop it. Similar to this, Islam et al. predicted the risk of diabetes in 520 people with 99% accuracy using a variety of machine learning techniques, such as Naive Bayes, Logistic Regression, and Random Forest. Some of the most recent techniques for RNA sequencing include deep learning via neural networks, autoencoders for data reduction, natural language processing for text mining, annotation for functional prediction, reinforcement learning for learning from new data, and adaptive learning algorithms for better diagnostic models through feedback from new data. Continuous research and technological advancements are gradually creating a path towards the effective application of AI-based RNA diagnostics in the clinical setting. We anticipate that these integrative developments will transform diabetes care in the future, thereby improving the quality of life for patients worldwide.
Challenges and future directions
RNA-based diagnostic tools are an untapped goldmine in the battle against diabetes, with specific and sensitive biomarkers related to the disease. However, there are several conditions that bar the application of such technologies in clinical cases, despite all the developments.
Some of the technical factors that impact RNA-based diagnostics include the stability of the RNA, the sensitivity of detection, sample processing, standardisation and reproducibility, data analysis and interpretation, patents, and commercialization barriers. Since RNA molecules are relatively unstable, they are easily degraded by enzymes called ribonucleases; therefore, their detection is slightly complicated. We need stable RNA procedures and various chemical modifications to overcome these challenges. Sample preparation is also another time-consuming stage; therefore, there is an essential need to develop automated extraction kits. Standard and good working practices have to be used to enable punctual execution, consistency, and reproducibility. Clinical validation is required as most of the RNA biomarkers have not been through the validation processes. Data collection and analysis are crucial and can only be done through the use of sophisticated bioinformatics programmes and skills. Approval is typically stringent because the authorities demand validation and documentation of the intended solution. Another challenge they face is the cost and availability of these solutions, as many are expensive and therefore not easily accessible to most people. Examples of solutions include extending efforts to rationally design cost-efficient diagnostic platforms and identifying potential reimbursement models to make RNA-based diagnostics more accessible.
The future of RNA-based diagnostics will witness advancements through the integration of nanotechnology, microfluidics, CRISPR-cas systems, and cross-disciplinary approaches. New approaches in nanotechnology help to increase the sensitivity and specificity of RNA detection, while microfluidics helps to reduce the cost of various diagnostic tools, making them more affordable. CRISPR-Cas systems include precision detection and versatile platforms. Personalised medicine includes individual treatment, prognosis, and long-term surveillance with the help of wearable technology. Real-time data can help make real-time changes to treatment regimens. There is a need for interdisciplinary collaboration mechanisms that can include research consortia, public-private partnerships, open source solutions, and social networking. These improvements will not only revolutionize the diagnostics field, but they will also accelerate the progression of RNA-based diagnostic commercialization. By investing in more research, RNA-based diagnostics can make diabetes care more effective and prompt, thus resulting in better healthcare for individuals with the disease.
The way forward
As indicated in Figure 1, the roadmap for enhancing RNA-based diagnostic tools in diabetes management is as follows:
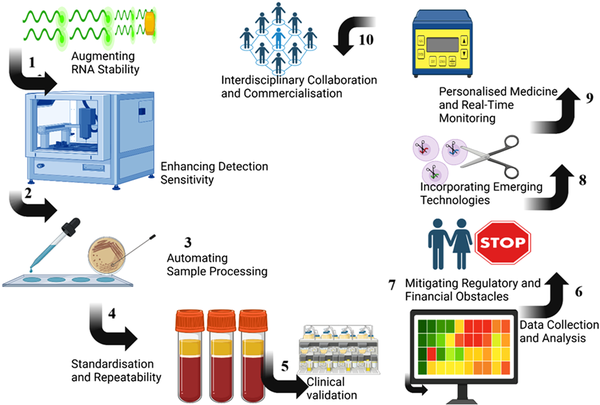
Figure 1
Flowchart/tiered roadmap diagram structure in enhancing RNA-based diagnostics in diabetes management.
1. Augmenting RNA Stability: RNA molecules are often susceptible to cleavage by ribonucleases, hence complicating detection. To address this issue, stable RNA methodologies must be developed, using various chemical modifications to enhance RNA stability.
2. Enhancing Detection Sensitivity: RNA-based biomarkers, often found in low quantities in blood, can be more accurately identified using advanced nanoscale technologies.
3. Automating Sample Processing: Sample preparation is one of the most time-consuming stages in RNA diagnostics. It would be prudent to develop automated extraction kits for sample extraction, hence minimising labour and time expenditure.
4. Standardisation and repeatability: It is advantageous to continually underscore that adherence to standard and exemplary laboratory methods enhances repeatability. This is essential, especially for the appropriate clinical use of RNA biomarkers.
5. Clinical Validation: Certain RNA biomarkers have not undergone consistent clinical verification. For RNA-based diagnostic assays to transition to clinical practice, validation stages will likely be necessary.
6. Data Collection and Analysis: Large-scale RNA data analysis need sophisticated bioinformatics tools and procedures to effectively interpret the data. Enhanced data management and the use of advanced software and machine learning algorithms will be utilised to improve data analysis and comprehension.
7. Mitigating Regulatory and Financial Obstacles: Regulators need extensive validation and documentation. Facilitating engagement with regulatory agencies to optimise approval processes and assess cost-effective diagnostic frameworks may improve the accessibility of RNA-based diagnostics.
8. Incorporating Emerging Technologies: The creation of tiny RNA-based diagnostic tools should integrate advanced technologies such as nanotechnology, microfluidics, and CRISPR-Cas systems. These developments will improve detection accuracy and simultaneously lower costs, with the development of multipurpose diagnostic tools.
9. Personalised Medicine and Real-Time Monitoring: The use of RNA in diagnostics facilitates the development of tailored medical treatments that align precisely with an individual’s genetic profile. Wearable technology allows for real-time data provision to facilitate the dynamic adjustment of treatment regimens based on individual factors.
10. Interdisciplinary Collaboration and Commercialisation: Commercialisation obstacles may be mitigated by multidisciplinary involvement, including robust public-private partnerships and consistent open-source initiatives. Securing financing for future research endeavours and collaboration with specialists in related disciplines will augment the use of RNA diagnostics in diabetes.
Conclusion
Diagnostics based on RNA are a major breakthrough in the diagnosis and monitoring of diabetes mellitus; they may shape the future development of diabetes care. These diagnostic tools would help to diagnose diabetes and its progression at an early stage and set specific treatment plans based on the molecular profiles of every patient due to the specificity and sensitivity of RNA biomarkers.
There are, however, some barriers to the use of RNA-based diagnostics in clinical practice. These include, but are not limited to, high RNA biomarker validation, establishing a proper method for diagnosing, and dealing with issues such as cost, availability, and implementation in current healthcare systems. However, RNA-based diagnostics have enormous potential for delivering more effective and efficient care to patients by enabling more accurate diagnoses. Further research to evaluate and validate the use of RNA based diagnostics in standard clinical practice is needed. Clinical and non-clinical collaboration, as well as that between industry partners, will be critical in overcoming the barriers to the use of RNA-based diagnostics and realizing its full potential. Therefore, enhancing these effective tools from the laboratory to the clinic through future research and technological advancements would improve diabetes treatment and patient care.
Author contributions Conceptualization: Esther Ugo Alum. Methodology: Esther Ugo Alum, Christian Emeka Offor, Ikechuku Okorie Igwenyi. Resources: Udu Ama Ibiam, Chris U. A. Ukaidi. Supervision: Obaroh Israel Olusegun, Christian Emeka Offor. Validation: Udu Ama Ibiam, Chris U. A. Ukaidi. Visualization: Ikechuku Okorie Igwenyi. Writing – original draft: Esther Ugo Alum, Ernest Nnamdi Ikpozu. Writing – review & editing: Esther Ugo Alum, Ernest Nnamdi Ikpozu, Christian Emeka Offor, Ikechuku Okorie Igwenyi, Obaroh Israel Olusegun, Udu Ama Ibiam, Chris U. A. Ukaidi.
Conflicting interests The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Mishra S, Tiwari P, Yadav R, et al. An extensive analysis of diseases associated with diabetes: review article. J Pharma Insights Res 2024; 2(3): 174–187.
- 2. Yedjou CG, Grigsby J, Mbemi A, et al. The management of diabetes mellitus using medicinal plants and vitamins. Int J Mol Sci 2023; 24(10): 9085.
- 3. Singh R, Gholipourmalekabadi M, Shafikhani SH. Animal models for type 1 and type 2 diabetes: advantages and limitations. Front Endocrinol 2024; 15: 1359685.
- 4. Patil SR, Chavan AB, Patel AM, et al. A review on diabetes mellitus its types, pathophysiology, epidermiology and its global burden. J Res Appl Sci Biotechnol 2023; 2(4): 73–79.
- 5. Alum E, Ugwu PC, Obeagu E. Beyond pregnancy: understanding the long-term implications of gestational diabetes mellitus. INOSR Sci Res 2024; 11: 63–71.
- 6. Aja P, Igwenyi I, Ugwu PC, et al. Evaluation of anti-diabetic effect and liver function indices of ethanol extracts of moringa oleifera and Cajanus cajan leaves in alloxan induced diabetic albino rats. 2015 1;439–447.
- 7. Abdalla MMI. Role of visfatin in obesity-induced insulin resistance. World J Clin Cases 2022; 10(30): 10840–10851.
- 8. Uti DE, Atangwho IJ, Eyong EU, et al. African walnuts (tetracarpidium conophorum) modulate hepatic lipid accumulation in obesity via reciprocal actions on HMG-CoA reductase and paraoxonase. Endocr, Metab Immune Disord: Drug Targets 2020; 20(3): 365–379.
- 9. Kreienkamp RJ, Voight BF, Gloyn AL, et al. Genetics of type 2 diabetes. In: Lawrence JM, Casagrande SS, Herman WH, et al. (eds) Diabetes in America. National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), 2023. https://www.ncbi.nlm.nih.gov/books/NBK597726/
- 10. Laakso M, Fernandes Silva L. Genetics of type 2 diabetes: past, present, and future. Nutrients 2022; 14(15): 3201.
- 11. Chawla G, Pradhan T, Gupta O. An insight into the combat strategies for the treatment of type 2 DiabetesMellitus. Mini Rev Med Chem 2024; 24(4): 403–430.
- 12. Nabi-Afjadi M, Ostadhadi S, Liaghat M, et al. Revolutionizing type 1 diabetes management: exploring oral insulin and adjunctive treatments. Biomed Pharmacother 2024; 176: 116808.
- 13. Obeagu EI, Ugwu OPC, Alum U. Poor glycaemic control among diabetic patients: a review on associated factors. Newport International Journal of Research in Medical Sciences. 2023; 3(1): 30–33.
- 14. Alum EU, Krishnamoorthy R, Gatasheh MK, et al. Protective role of jimson weed in mitigating dyslipidemia, cardiovascular, and renal dysfunction in diabetic rat models: in vivo and in silico evidence. Nat Prod Commun 2024; 19(12): 1934578X241299279.
- 15. International Diabetes Federation. Facts & figures. Available from: https://idf.org/about-diabetes/diabetes-facts-figures/
- 16. Ugwu OPS, Kungu E, Inyangat R, et al. Exploring Indigenous Medicinal Plants for Managing Diabetes Mellitus in Uganda: Ethnobotanical Insights, Pharmacotherapeutic Strategies, and National Development Alignment. INOSR Experimental Sciences 2023; 12(2): 214–224. https://doi.org/10.59298/INOSRES/2023/2.17.1000.
- 17. Alum E, Ugwu PC, Obeagu E, et al. Managing the dual burden: addressing mental health in. Diabetes Care 2024; 2: 1–9.
- 18. Damase TR, Sukhovershin R, Boada C, et al. The limitless future of RNA therapeutics. Front Bioeng Biotechnol 2021; 9: 628137. https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2021.628137/full
- 19. Dehghanbanadaki H, Asili P, Haji Ghadery A, et al. Diagnostic accuracy of circular RNA for diabetes mellitus: a systematic review and diagnostic meta-analysis. BMC Med Genom 2023; 16: 48.
- 20. Man HSJ, Moosa VA, Singh A, et al. Unlocking the potential of RNA-based therapeutics in the lung: current status and future directions. Front Genet 2023; 14: 1281538. https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2023.1281538/full
- 21. López-Urrutia E, Bustamante Montes LP, Ladrón de Guevara Cervantes D, et al. Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front Oncol 2019; 9: 669. https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2019.00669/full
- 22. Boroumand F, Saadat I, Saadat M. Non-randomness distribution of micro-RNAs on human chromosomes. Egypt J Med Hum Genet 2019; 20(1): 33.
- 23. O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol 2018; 9: 402.
- 24. Vasu S, Kumano K, Darden CM, et al. MicroRNA signatures as future biomarkers for diagnosis of diabetes states. Cells 2019; 8(12): 1533.
- 25. Mishra S, Bahinipati J, Sarangi R, et al. A comprehensive overview on micro RNA signature in type 2 diabetes mellitus and its complications. Indian J Clin Biochem 2023; 38(2): 151–158.
- 26. Kim M, Zhang X. The profiling and role of miRNAs in diabetes mellitus. J Diabetes Clin Res 2019; 1(1): 5–23.
- 27. Arevalo-Martinez M, Cidad P, Moreno-Estar S, et al. miR-126 contributes to the epigenetic signature of diabetic vascular smooth muscle and enhances antirestenosis effects of Kv1.3 blockers. Mol Metabol 2021; 53: 101306.
- 28. Sidorkiewicz I, Niemira M, Maliszewska K, et al. Circulating miRNAs as a predictive biomarker of the progression from prediabetes to diabetes: outcomes of a 5-year prospective observational study. J Clin Med 2020; 9(7): 2184.
- 29. Kamalden TA, Macgregor-Das AM, Kannan SM, et al. Exosomal MicroRNA-15a transfer from the pancreas augments diabetic complications by inducing oxidative stress. Antioxidants Redox Signal 2017; 27(13): 913–930.
- 30. Jiménez-Lucena R, Rangel-Zúñiga OA, Alcalá-Díaz JF, et al. Circulating miRNAs as predictive biomarkers of type 2 diabetes mellitus development in coronary heart disease patients from the CORDIOPREV study. Mol Ther Nucleic Acids 2018; 12: 146–157.
- 31. Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021; 22(2): 96–118.
- 32. Dieter C, Lemos NE, Corrêa NRF, et al. The impact of lncRNAs in diabetes mellitus: a systematic review and in silico analyses. Front Endocrinol 2021; 12: 602597.
- 33. Wu Q, Huang F. LncRNA H19: a novel player in the regulation of diabetic kidney disease. Front Endocrinol 2023; 14: 1238981. https://www.frontiersin.org/journals/endocrinology/articles/10.3389/fendo.2023.1238981/full
- 34. Fan W, Peng Y, Liang Z, et al. A negative feedback loop of H19/miR-675/EGR1 is involved in diabetic nephropathy by downregulating the expression of the vitamin D receptor. J Cell Physiol 2019; 234(10): 17505–17513.
- 35. Che F, Han Y, Fu J, et al. LncRNA MALAT1 induced by hyperglycemia promotes microvascular endothelial cell apoptosis through activation of the miR-7641/TPR axis to exacerbate neurologic damage caused by cerebral small vessel disease. Ann Transl Med 2021; 9(24): 1762.
- 36. Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 2016; 12(6): 360–373.
- 37. Gong W, Zhu G, Li J, et al. LncRNA MALAT1 promotes the apoptosis and oxidative stress of human lens epithelial cells via p38MAPK pathway in diabetic cataract. Diabetes Res Clin Pract 2018; 144: 314–321.
- 38. Geng M, Liu W, Li J, et al. LncRNA as a regulator in the development of diabetic complications. Front Endocrinol 2024; 15: 1324393.
- 39. Yang Q, Fang D, Chen J, et al. LncRNAs associated with oxidative stress in diabetic wound healing: regulatory mechanisms and application prospects. Theranostics 2023; 13(11): 3655–3674.
- 40. Yang Z, Xu F, Teschendorff AE, et al. Insights into the role of long non-coding RNAs in DNA methylation mediated transcriptional regulation. Front Mol Biosci 2022; 9: 1067406.
- 41. Mattick JS, Amaral PP, Carninci P, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 2023; 24(6): 430–447.
- 42. Xiao J, Joseph S, Xia M, et al. Circular RNAs acting as miRNAs’ sponges and their roles in stem cells. J Clin Med 2022; 11(10): 2909.
- 43. Yin W, Zhang Z, Xiao Z, et al. Circular RNAs in diabetes and its complications: current knowledge and future prospects. Front Genet 2022; 13: 1006307.
- 44. Liu M, Zhao J. Circular RNAs in diabetic nephropathy: updates and perspectives. Aging Dis 2022; 13(5): 1365–1380.
- 45. Stoll L, Rodríguez-Trejo A, Guay C, et al. A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat Commun 2020; 11: 5611.
- 46. Brozzi F, Regazzi R. Circular RNAs as novel regulators of β-cell functions under physiological and pathological conditions. Int J Mol Sci 2021; 22(4): 1503.
- 47. Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017; 136(17): 1629–1642.
- 48. Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metabol 2018; 9: 69–83.
- 49. Feher J. 2.2 - DNA and protein synthesis. In: Feher J (ed) Quantitative human physiology (2nd ed.) Academic Press; 2017, pp. 120–129. https://www.sciencedirect.com/science/article/pii/B9780128008836000112
- 50. Qin S, Tang X, Chen Y, et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct Targeted Ther 2022; 7: 166.
- 51. Bozadjieva Kramer N, Lubaczeuski C, Blandino-Rosano M, et al. Glucagon resistance and decreased susceptibility to diabetes in a model of chronic hyperglucagonemia. Diabetes 2021; 70(2): 477–491.
- 52. Finan B, Capozzi ME, Campbell JE. Repositioning glucagon action in the physiology and pharmacology of diabetes. Diabetes 2020; 69(4): 532–541.
- 53. Suomi T, Starskaia I, Kalim UU, et al. Gene expression signature predicts rate of type 1 diabetes progression. EBioMedicine 2023; 92: 104625.
- 54. Di Bella MA. Overview and update on extracellular vesicles: considerations on exosomes and their application in modern medicine. Biology 2022; 11(6): 804.
- 55. Wang J, Yue BL, Huang YZ, et al. Exosomal RNAs: novel potential biomarkers for diseases—a review. Int J Mol Sci 2022; 23(5): 2461.
- 56. González-Blanco C, Iglesias-Fortes S, Lockwood ÁC, et al. The role of extracellular vesicles in metabolic diseases. Biomedicines 2024; 12(5): 992.
- 57. Ashrafizadeh M, Kumar AP, Aref AR, et al. Exosomes as promising nanostructures in diabetes mellitus: from insulin sensitivity to ameliorating diabetic complications. Int J Nanomed 2022; 17: 1229–1253.
- 58. Sun Y, Shi H, Yin S, et al. Human mesenchymal stem cell derived exosomes alleviate type 2 diabetes mellitus by reversing peripheral insulin resistance and relieving β-cell destruction. ACS Nano 2018; 12(8): 7613–7628.
- 59. Sun Y, Mao Q, Shen C, et al. Exosomes from β-cells alleviated hyperglycemia and enhanced angiogenesis in islets of streptozotocin-induced diabetic mice. Diabetes Metab Syndr Obes 2019; 12: 2053–2064.
- 60. Jovic D, Liang X, Zeng H, et al. Single‐cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med 2022; 12(3): e694.
- 61. Wang S, Sun ST, Zhang XY, et al. The evolution of single-cell RNA sequencing technology and application: progress and perspectives. Int J Mol Sci 2023; 24(3): 2943.
- 62. Fu Q, Jiang H, Qian Y, et al. Single-cell RNA sequencing combined with single-cell proteomics identifies the metabolic adaptation of islet cell subpopulations to high-fat diet in mice. Diabetologia 2023; 66(4): 724–740.
- 63. Huang D, Ma N, Li X, et al. Advances in single-cell RNA sequencing and its applications in cancer research. J Hematol Oncol 2023; 16: 98.
- 64. Reid AJ, Talman AM, Bennett HM, et al. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 2018; 7: e33105.
- 65. Ahmed R, Zaman T, Chowdhury F, et al. Single-cell RNA sequencing with spatial transcriptomics of cancer tissues. Int J Mol Sci 2022; 23(6): 3042.
- 66. Li J, Zheng Y, Yan P, et al. A single-cell transcriptomic atlas of primate pancreatic islet aging. Natl Sci Rev 2020; 8(2): nwaa127.
- 67. Li Y, Liu Y, Liu S, et al. Diabetic vascular diseases: molecular mechanisms and therapeutic strategies. Signal Transduct Targeted Ther 2023; 8(1): 152–229.
- 68. Chen K, Zhang J, Huang Y, et al. Single-cell RNA-seq transcriptomic landscape of human and mouse islets and pathological alterations of diabetes. iScience 2022; 25(11): 105366.
- 69. Song Y, He C, Jiang Y, et al. Bulk and single-cell transcriptome analyses of islet tissue unravel gene signatures associated with pyroptosis and immune infiltration in type 2 diabetes. Front Endocrinol 2023; 14: 1132194.
- 70. Ma L, Zheng J. Single-cell gene expression analysis reveals β-cell dysfunction and deficit mechanisms in type 2 diabetes. BMC Bioinf 2018; 19(Suppl 19): 515.
- 71. Augsornworawat P, Millman JR. Single-cell RNA sequencing for engineering and studying human islets. Curr Opin Biomed Eng 2020; 16: 27–33.
- 72. Ji L, Guo W. Single-cell RNA sequencing highlights the roles of C1QB and NKG7 in the pancreatic islet immune microenvironment in type 1 diabetes mellitus. Pharmacol Res 2023; 187: 106588.
- 73. Muñoz García A, Juksar J, Groen N, et al. Single-cell transcriptomics reveals a role for pancreatic duct cells as potential mediators of inflammation in diabetes mellitus. Front Immunol 2024; 15: 1381319. https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1381319/full
- 74. Kang H, Lee J. Adipose tissue macrophage heterogeneity in the single-cell genomics era. Mol Cells 2024; 47(2): 100031.
- 75. Henriques F, Bedard AH, Guilherme A, et al. Single-cell RNA profiling reveals adipocyte to macrophage signaling sufficient to enhance thermogenesis. Cell Rep 2020; 32(5): 107998.
- 76. He L, Lu A, Qin L, et al. Application of single-cell RNA sequencing technology in liver diseases: a narrative review. Ann Transl Med 2021; 9(20): 1598.
- 77. Atif J, Thoeni C, Bader GD, et al. Unraveling the complexity of liver disease one cell at a time. Semin Liver Dis 2022; 42(3): 250–270.
- 78. Yang T, Yan C, Yang L, et al. Identification and validation of core genes for type 2 diabetes mellitus by integrated analysis of single-cell and bulk RNA-sequencing. Eur J Med Res 2023; 28(1): 340.
- 79. Zhang X, Liu L. Applications of single cell RNA sequencing to research of stem cells. World J Stem Cell 2019; 11(10): 722–728.
- 80. Li H, Wang Y, Zhu G, et al. Application progress of single-cell sequencing technology in mesenchymal stem cells research. Front Cell Dev Biol 2024; 11: 1336482. https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2023.1336482/full
- 81. Naydenov DD, Vashukova ES, Barbitoff YA, et al. Current status and prospects of the single-cell sequencing technologies for revealing the pathogenesis of pregnancy-associated disorders. Genes 2023; 14(3): 756.
- 82. Moyce BL, Dolinsky VW. Maternal β-cell adaptations in pregnancy and placental signalling: implications for gestational diabetes. Int J Mol Sci 2018; 19(11): 3467.
- 83. Zhang X, Zhang F, Xu X. Single‐cell RNA sequencing in exploring the pathogenesis of diabetic retinopathy. Clin Transl Med 2024; 14(7): e1751. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC11214886/
- 84. Zhang R, Huang C, Chen Y, et al. Single-cell transcriptomic analysis revealing changes in retinal cell subpopulation levels and the pathways involved in diabetic retinopathy. Ann Transl Med 2022; 10(10): 562. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9201190/
- 85. Hanna SJ, Tatovic D, Thayer TC, et al. Insights from single cell RNA sequencing into the immunology of type 1 diabetes- cell phenotypes and antigen specificity. Front Immunol 2021; 12: 751701.
- 86. Wang W, Zhang C. Targeting β-cell dedifferentiation and transdifferentiation: opportunities and challenges. Endocr Connect 2021; 10(8): R213–R228.
- 87. Khin PP, Lee JH, Jun HS. A brief review of the mechanisms of β-cell dedifferentiation in type 2 diabetes. Nutrients 2021; 13(5): 1593.
- 88. Fu J, Akat KM, Sun Z, et al. Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J Am Soc Nephrol 2019; 30(4): 533–545.
- 89. Xu Y, Xiang Z, E W, et al. Single-cell transcriptomes reveal a molecular link between diabetic kidney and retinal lesions. Commun Biol 2023; 6(1): 912.
- 90. Cheng C, Chen W, Jin H, et al. A review of single-cell RNA-seq annotation, integration, and cell–cell communication. Cells 2023; 12(15): 1970.
- 91. Gondal MN, Shah SUR, Chinnaiyan AM, et al. A systematic overview of single-cell transcriptomics databases, their use cases, and limitations. Front Bioinform 2024; 4: 1417428.
- 92. Leote AC, Wu X, Beyer A. Regulatory network-based imputation of dropouts in single-cell RNA sequencing data. PLoS Comput Biol 2022; 18(2): e1009849.
- 93. Margaritis K, Margioula-Siarkou G, Giza S, et al. Micro-RNA implications in type-1 diabetes mellitus: a review of literature. Int J Mol Sci 2021; 22(22): 12165.
- 94. Cheruiyot A, Hollister-Lock J, Sullivan BA, et al. Sustained hyperglycemia specifically targets translation of mRNAs for insulin secretion. J Clin Invest. 2023. https://www.jci.org/articles/view/173280
- 95. Al-Mahayni S, Ali M, Khan M, et al. Glycemia-induced miRNA changes: A review. Int J Mol Sci. 2023; 24(8): 7488. doi:10.3390/ijms24087488.
- 96. Firdos PT, Pramanik T, Verma P, et al. (Re-)viewing role of intracellular glucose beyond extracellular regulation of glucose-stimulated insulin secretion by pancreatic cells. ACS Omega 2024; 9(10): 11755–11768.
- 97. Grieco GE, Brusco N, Licata G, et al. The landscape of microRNAs in βCell: between phenotype maintenance and protection. Int J Mol Sci 2021; 22(2): 803.
- 98. Tello-Flores VA, Beltrán-Anaya FO, Ramírez-Vargas MA, et al. Role of long non-coding RNAs and the molecular mechanisms involved in insulin resistance. Int J Mol Sci 2021; 22(14): 7256.
- 99. Yang W, Lyu Y, Xiang R, et al. Long noncoding RNAs in the pathogenesis of insulin resistance. Int J Mol Sci 2022; 23(24): 16054.
- 100. Hwang HJ, Kim YK. Molecular mechanisms of circular RNA translation. Exp Mol Med 2024; 56: 1272–1280.
- 101. Li Z, Ren Y, Lv Z, et al. Decrypting the circular RNAs does a favor for us: understanding, diagnosing and treating diabetes mellitus and its complications. Biomed Pharmacother 2023; 168: 115744.
- 102. Dailey G. Assessing glycemic control with self-monitoring of blood glucose and hemoglobin A(1c) measurements. Mayo Clin Proc 2007; 82: 229–235. https://www.semanticscholar.org/paper/Assessing-glycemic-control-with-self-monitoring-of-Dailey/4977b06f93262bf4b85eb307e9d9981435c53ef3
- 103. Fathima J, Anudeepa J, Muralidharan P, et al. A review on continuous glucose monitoring in diabetes management. In: Proceedings of the first international conference on science, engineering and technology practices for sustainable development ICSETPSD 2023, Coimbatore, India, 17th-18th November 2023, 2024. https://eudl.eu/doi/10.4108/eai.17-11-2023.2342847
- 104. Rodbard D. Continuous glucose monitoring: a review of recent studies demonstrating improved glycemic outcomes. Diabetes Technol Therapeut 2017; 19(S3): S25–S37.
- 105. Raja JM, Maturana MA, Kayali S, et al. Diabetic foot ulcer: a comprehensive review of pathophysiology and management modalities. World J Clin Cases 2023; 11(8): 1684–1693.
- 106. Zhang Y, Zhang J, Xu Z, et al. Regulation of NcRNA-protein binding in diabetic foot. Biomed Pharmacother 2023; 160: 114361.
- 107. Chao HM, Wang TW, Chern E, et al. Regulatory RNAs, microRNA, long-non coding RNA and circular RNA roles in colorectal cancer stem cells. World J Gastrointest Oncol 2022; 14(4): 748–764.
- 108. Sakshi S, Jayasuriya R, Sathish Kumar RC, et al. MicroRNA-27b impairs nrf2-mediated angiogenesis in the progression of diabetic foot ulcer. J Clin Med 2023; 12(13): 4551.
- 109. Deng H, Li B, Shen Q, et al. Mechanisms of diabetic foot ulceration: a review. J Diabetes 2023; 15(4): 299–312.
- 110. Lyttle BD, Vaughn AE, Bardill JR, et al. Effects of microRNAs on angiogenesis in diabetic wounds. Front Med 2023; 10: 1140979.
- 111. Ma L, Wen Y, Li Z, et al. Circulating MicroRNAs as potential diagnostic biomarkers for diabetic retinopathy: a meta-analysis. Front Endocrinol 2022; 13: 929924.
- 112. Juárez-Vicuña Y, Ruiz-Ojeda D, González-Ramírez J, et al. LncRNA MALAT1 in Keratinocyte function: a review of recent advances. Noncoding RNA Res 2024; 9(2): 594–601.
- 113. Yan C, Chen J, Yang X, et al. Emerging roles of long non-coding RNAs in diabetic foot ulcers. Diabetes Metab Syndr Obes 2021; 14: 2549–2560.
- 114. Loganathan T, Doss C GP. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genomics 2023; 23(1): 33.
- 115. Ikeda Y, Morikawa S, Nakashima M, et al. CircRNAs and RNA-binding proteins involved in the pathogenesis of cancers or central nervous system disorders. Noncoding RNA 2023; 9(2): 23.
- 116. Bhatt T, Dey R, Hegde A, et al. Initiation of wound healing is regulated by the convergence of mechanical and epigenetic cues. PLoS Biol 2022; 20(9): e3001777.
- 117. Wang Y, Shao T, Wang J, et al. An update on potential biomarkers for diagnosing diabetic foot ulcer at early stage. Biomed Pharmacother 2021; 133: 110991.
- 118. Chen P, Pang G, Cai F, et al. Strain improvement and genetic engineering of trichoderma for industrial applications. In: Zaragoza Ó, Casadevall A (eds) Encyclopedia of mycology. Elsevier, 2021, pp. 505–517. https://www.sciencedirect.com/science/article/pii/B9780128199909000299
- 119. Clark DP, Pazdernik NJ. Chapter 18 - regulation at the RNA level. In: Clark DP, Pazdernik NJ (eds) Molecular Biology (2nd ed.) Academic Press, 2013, pp. 553–580. https://www.sciencedirect.com/science/article/pii/B9780123785947000184
- 120. Hu B, Zhong L, Weng Y, et al. Therapeutic siRNA: state of the art. Signal Transduct Targeted Ther 2020; 5(1): 101–125.
- 121. Ahmed SAH, Ansari SA, Mensah-Brown EPK, et al. The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin Epigenet 2020; 12(1): 104.
- 122. Isenmann M, Stoddart MJ, Schmelzeisen R, et al. Basic principles of RNA interference: nucleic acid types and in vitro intracellular delivery methods. Micromachines 2023; 14(7): 1321.
- 123. Roy P, Saha S, Chakrabarty J. Looking into the possibilities of cure of the type 2 diabetes mellitus by nanoparticle-based RNAi and CRISPR-Cas9 system: a review. J Drug Deliv Sci Technol 2021; 66: 102830.
- 124. Campbell JE, Newgard CB. Mechanisms controlling pancreatic islet cell function in insulin secretion. Nat Rev Mol Cell Biol 2021; 22(2): 142–158.
- 125. Aghaei-Zarch SM. Crosstalk between MiRNAs/lncRNAs and PI3K/AKT signaling pathway in diabetes mellitus: mechanistic and therapeutic perspectives. Noncoding RNA Res 2024; 9(2): 486–507.
- 126. Hu K, Liu L, Tang S, et al. MicroRNA-221-3p inhibits the inflammatory response of keratinocytes by regulating the DYRK1A/STAT3 signaling pathway to promote wound healing in diabetes. Commun Biol 2024; 7(1): 300–315.
- 127. Friedrich M, Aigner A. Therapeutic siRNA: state-of-the-art and future perspectives. BioDrugs 2022; 36(5): 549–571.
- 128. Zhang J, Chen B, Gan C, et al. A comprehensive review of small interfering RNAs (siRNAs): mechanism, therapeutic targets, and delivery strategies for cancer therapy. Int J Nanomed 2023; 18: 7605–7635.
- 129. Bhatia D, Paul S, Acharjee T, et al. Biosensors and their widespread impact on human health. Sens Int 2024; 5: 100257.
- 130. Yeung AWK, Torkamani A, Butte AJ, et al. The promise of digital healthcare technologies. Front Public Health 2023; 11: 1196596.
- 131. Haleem A, Javaid M, Singh RP, et al. Telemedicine for healthcare: capabilities, features, barriers, and applications. Sens Int 2021; 2: 100117.
- 132. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, et al. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 2016; 17(5): 257–271.
- 133. Madrid RE, Ashur Ramallo F, Barraza DE, et al. Smartphone-based biosensor devices for healthcare: technologies, trends, and adoption by end-users. Bioengineering 2022; 9(3): 101.
- 134. Ezenwaji CO, Alum EU, Ugwu OPC. The role of digital health in pandemic preparedness and response: securing global health? Glob Health Action 2024; 17(1): 2419694.
- 135. Wu XF, Shen CF, Cheng CM. Integration of mobile devices and point-of-care diagnostic devices—the case of C-reactive protein diagnosis. Diagnostics 2019; 9(4): 181.
- 136. Prabhakar G, Chintala VR, Reddy T, et al. User-cloud-based ensemble framework for type-2 diabetes prediction with diet plan suggestion. E-Prime - Adv Electr Eng Electron Energy 2024; 7: 100423.
- 137. Singh SU, Chatterjee S, Lone SA, et al. Advanced wearable biosensors for the detection of body fluids and exhaled breath by graphene. Mikrochim Acta 2022; 189(6): 236.
- 138. Gao Q, Li S. Intelligent point of care testing for medicine diagnosis. Interdiscip Med 2024; 2(1): e20230031.
- 139. Wang J, Davidson JL, Kaur S, et al. Paper-based biosensors for the detection of nucleic acids from pathogens. Biosensors 2022; 12(12): 1094.
- 140. Angelescu MA, Andronic O, Dima SO, et al. miRNAs as biomarkers in diabetes: moving towards precision medicine. Int J Mol Sci 2022; 23(21): 12843.
- 141. Faura G, Boix-Lemonche G, Holmeide AK, et al. Colorimetric and electrochemical screening for early detection of diabetes mellitus and diabetic retinopathy—application of sensor arrays and machine learning. Sensors 2022; 22(3): 718.
- 142. Lu T, Ji S, Jin W, et al. Biocompatible and long-term monitoring strategies of wearable, ingestible and implantable biosensors: reform the next generation healthcare. Sensors 2023; 23(6): 2991.
- 143. Shoaib A, Darraj A, Khan ME, et al. A nanotechnology-based approach to biosensor application in current diabetes management practices. Nanomaterials 2023; 13(5): 867.
- 144. Sugandh FNU, Chandio M, Raveena FNU, et al. Advances in the management of diabetes mellitus: a focus on personalized medicine. Cureus 2023; 15(8): e43697. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10505357/
- 145. Karalis VD. The integration of artificial intelligence into clinical practice. Appl Biosci 2024; 3(1): 14–44.
- 146. Alowais SA, Alghamdi SS, Alsuhebany N, et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ 2023; 23(1): 689.
- 147. Brendel M, Su C, Bai Z, et al. Application of deep learning on single-cell rna sequencing data analysis: a review. Genom Proteom Bioinform 2022; 20(5): 814–835.
- 148. Sk Eal R. Machine learning and deep learning models for predicting the onset of diabetes: a pilot study. Int J Recent Innov Trends Comput Commun 2023; 11(9): 3564–3573.
- 149. Chen S, Guo W. Auto-encoders in deep learning—a review with new perspectives. Mathematics 2023; 11(8): 1777.
- 150. Chandra B, Sharma RK. Exploring autoencoders for unsupervised feature selection. In: 2015 international joint conference on neural networks, Killarney, 2015, pp. 1–6.
- 151. Pratella D, Ait-El-Mkadem Saadi S, Bannwarth S, et al. A survey of autoencoder algorithms to pave the diagnosis of rare diseases. Int J Mol Sci 2021; 22(19): 10891.
- 152. Oroojeni Mohammad Javad M, Agboola SO, Jethwani K, et al. A reinforcement learning–based method for management of type 1 diabetes: exploratory study. JMIR Diabetes 2019; 4(3): e12905.
- 153. Mansur A, Vrionis A, Charles JP, et al. The role of artificial intelligence in the detection and implementation of biomarkers for hepatocellular carcinoma: outlook and opportunities. Cancers 2023; 15(11): 2928.
- 154. Khalighi S, Reddy K, Midya A, et al. Artificial intelligence in neuro-oncology: advances and challenges in brain tumor diagnosis, prognosis, and precision treatment. npj Precis Oncol 2024; 8(1): 1–12.
- 155. Nguyen LP, Tung DD, Nguyen DT, et al. The utilization of machine learning algorithms for assisting physicians in the diagnosis of diabetes. Diagnostics 2023; 13(12): 2087.
- 156. Shukla AK. Patient diabetes forecasting based on machine learning approach. In: Pant M, Kumar Sharma T, Arya R, et al. (eds) Advances in intelligent systems and computing. Springer Singapore, 2020, Vol. 1154, pp. 1017–1027. https://link.springer.com/10.1007/978-981-15-4032-5_91
- 157. Islam MMF, Ferdousi R, Rahman S, et al. Likelihood prediction of diabetes at early stage using data mining techniques. In: Computer vision and machine intelligence in medical image analysis. Springer, 2020, pp. 113–125.
- 158. Erfanian N, Heydari AA, Feriz AM, et al. Deep learning applications in single-cell genomics and transcriptomics data analysis. Biomed Pharmacother 2023; 165: 115077.
- 159. Wang Y, Zhu D, Yu H, et al. Non-coding RNAs function as diagnostic biomarkers and therapeutic targets in Pulmonary arterial hypertension. IntechOpen, 2024. https://www.intechopen.com/online-first/1180982
- 160. Gradisteanu Pircalabioru G, Musat M, Elian V, et al. Liquid biopsy: a game changer for type 2 diabetes. Int J Mol Sci 2024; 25(5): 2661.
- 161. Puig-Serra P, Casado-Rosas MC, Martinez-Lage M, et al. CRISPR approaches for the diagnosis of human diseases. Int J Mol Sci 2022; 23(3): 1757.
- 162. Kumari M, Gupta V, Kumar N, et al. Microfluidics-based nanobiosensors for healthcare monitoring. Mol Biotechnol 2024; 66(3): 378–401.
- 163. Tyler J, Choi SW, Tewari M. Real-time, personalized medicine through wearable sensors and dynamic predictive modeling: a new paradigm for clinical medicine. Curr Opin Struct Biol 2020; 20: 17–25.
Abbreviations
RNA: Ribonucleic acid
RNAi: RNA interference
mRNA: messenger RNA
VIGS: Virus-induced gene silencing
DCLs: Dicer-like enzymes
dsRNA: Double-stranded RNA
siRNAs: Small interfering RNAs
RISC: RNA-induced silencing complex
T1DM: Type 1 diabetes mellitus
T2DM: Type 2 diabetes mellitus
HbA1c: Glycated hemoglobin.