Introduction
Salmonella enterica infections progress in intracellular locations of phagocytic and non-phagocytic host cells. The avidity of this pathogen to be rapidly internalized by host cells imposes a barrier for commonly used antibiotics, which are more effective in extracellular locations., Many studies have shown limited accessibility of drugs to the intracellular niche occupied by the pathogen, a factor decreasing their inhibitory activity. In intraphagosomal intracellular pathogens, other factors such the acidity of the compartment colonized by the pathogen can also affect antibiotic activity. To counterbalance these negative effects, new approaches based on highly penetrating nanoparticles carrying the drug as cargo are currently under intense investigation and development.
The clinical evidence accumulated in S. enterica infections both in livestock and humans indicates that this pathogen is prone to cause persistent infections., The reduced proliferation rate inside the infected cell, associated in many instances with latency, imposes an additional drawback for chemotherapy, which normally requires the pathogen to be undergoing active metabolism. The standard antimicrobial therapy based on cephalosporins and quinolones seems in some instances ineffective to eradicate S. enterica infections. In this scenario, it is alarming that relapse caused by drug-susceptible isolates is reported in the order of 5%–15% by many studies., Intracellularity as a lifestyle therefore impacts the antimicrobial chemotherapy of S. enterica infections.
Our recent studies show an additional factor hampering effective treatment of S. enterica infections. Besides PBP2 and PBP3, the two peptidoglycan synthases conserved in enteric bacteria that are responsible for cell elongation and division, respectively,S. enterica encodes two homologous enzymes named PBP2SAL and PBP3SAL, which replace PBP2 and PBP3 when located inside host cells. Intracellular S. enterica therefore upregulates certain functions compared with bacteria growing extracellularly, and among these are enzymes related to the metabolism of peptidoglycan, one of the main targets in antimicrobial therapy. Importantly, it has been shown in a mouse animal model that an S. Typhimurium mutant lacking PBP3SAL does not cause relapse following ceftriaxone therapy, highlighting how PBP3SAL contributes to prevent effective eradication of Salmonella infections. These observations indicate that new antimicrobial therapies directed to selectively inhibit PBP2SAL and/or PBP3SAL are imperative to control intracellular infections caused by this pathogen.
In this study, we identified the second-generation β-lactam cephalosporin cefotiam (IUPAC name: (6R,7R)-7-[[2-(2-amino-1,3-thiazol-4-yl)-acetyl]amino]-3-[[1-[2-(dimethylamino)ethyl]tetrazol-5-yl]sulfanylmethyl]-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid) as an antibiotic showing higher affinity for PBP3SAL than for PBP3. Consistently with the replacement of these two PBPs by intracellular S. Typhimurium, cefotiam proved to be effective in controlling the infection inside host cells in a PBP3SAL-dependent manner. Conversely, cefuroxime, a cephalosporin with higher affinity for PBP3 than for PBP3SAL, showed reduced effectiveness in the killing of intracellular bacteria.
Methods
Prestwick Chemical Library and selected compounds
The Prestwick Chemical Library (Prestwick Chemical Libraries, GreenPharma S.A.S., Orléans, France) was used to screen compounds that induce filamentation in the S. Typhimurium ΔPBP3 mutant having PBP3SAL as the only PBP promoting cell division. The library has 1520 off-patent compounds approved by agencies such as the FDA and the EMA. The compounds were supplied at 10 mM in 100% DMSO.
Bacterial strains, eukaryotic cell lines and growth conditions
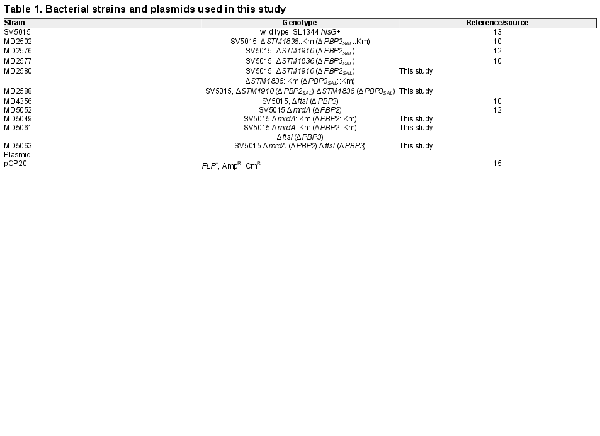
S. Typhimurium strains used were isogenic to the parental wild-type strain SV5015, a His+ derivative of the virulent strain SL1344 isolated from calf. The strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in LB broth buffered with 80 mM MES [2-(N-morpholino) ethanesulfonic acid] and adjusted to pH 4.6. When necessary, ampicillin and kanamycin were added at 0.1 mg/mL and 0.03 mg/mL, respectively. The S. Typhimurium double mutants MD5063 [ΔmrdA (ΔPBP2) ΔftsI (ΔPBP3)] and MD2588 [ΔSTM1910 (ΔPBP2SAL) ΔSTM1836 (ΔPBP3SAL)] were constructed by bacteriophage P22-mediated transduction using donor strains MD2502 [ΔSTM1836::Km (ΔPBP3SAL::Km)] and MD5049 [ΔmrdA::Km (ΔPBP2::Km)]. The P22 lysates obtained in MD2502 and MD5409 strains, were used to transduce recipient strains MD2576 [ΔSTM1910 (ΔPBP2SAL)] and MD4356 [ΔftsI (ΔPBP3)], respectively. Subsequent removal of the kanamycin cassette from strains MD2580 [ΔSTM1910 (ΔPBP2SAL) ΔSTM1836::Km (ΔPBP3SAL::Km)] and MD5061 [ΔmrdA::Km (ΔPBP2::Km) ΔftsI (ΔPBP3)] (see Table 1), was performed as described.
Identification of drugs blocking cell division based on monitoring of OD600 values
As a previous control, the growth of S. Typhimurium wild-type and ΔPBP3 isogenic strains was monitored in LB medium at pH 4.6 in the absence/presence of 0.001 mg/mL aztreonam using a Spark Automatic Microplate Reader (Tecan Trading AG, Switzerland). Differences in final OD600 values were correlated with the presence of filamented bacteria in which cell division was blocked by the antibiotic. Response to compounds of the Prestwick Library was directed to the monitoring of growth curves and final OD600 values. Drugs having a bacteriostatic effect were discarded from further analyses. Those compounds causing bacteriolytic effect at the initial dose used (1:100 dilution of the 10 mM stock) were rescreened at lower concentrations and reanalysed for increased values in the final OD600 measurements. Growth curves were monitored in the automatic reader for a minimum period of 8 h. Control samples consisted of bacteria grown in LB medium pH 4.6 containing 1% DMSO.
Antibiotic susceptibility tests
MIC values were determined using MIC strips (MIC Test Strip, Liofilchem, Roseto degli Abruzzi, Italy) in LB plates at pH 4.6. Because no cefotiam MIC strips are commercially available, MIC values for this antibiotic were determined in 96-well microplates by serial dilutions of the antibiotic in liquid LB pH 4.6 medium and overnight incubation at 37°C. The starting inoculum was 6 × 105 cfu per well.
Preparation of membrane extracts for BOCILLIN-FL (Boc-FL) labelling
S. Typhimurium ΔftsI (ΔPBP3) and ΔSTM1836 (ΔPBP3SAL) isogenic strains were grown overnight at 37°C in LB pH 4.6. Cultures were diluted 1:100 in 200 mL fresh media and bacteria grown up to exponential phase (OD600 ∼0.2–0.3). Bacteria were harvested by centrifugation (6000×g, 10 min, 4°C) and washed in 50 mM sodium phosphate buffer pH 4.6. After subsequent centrifugation (12 000×g, 15 min, 4°C), bacteria were suspended in 20 mL of 50 mM sodium phosphate buffer. Cells were disrupted by passing through a French press and lysates centrifuged at low speed (4000×g, 10 min, 4°C) to remove unbroken cells. The supernatant was further centrifuged (150 000×g, 35 min, 4°C) and pellets containing membrane material were suspended in 150 μL of 50 mM sodium phosphate buffer pH 4.6. The protein concentration was measured using Pierce 660 nm Protein Assay reagent (Thermo Scientific).
Cefotiam competition in Boc-FL binding assays
Cefotiam hydrochloride (ref. 66309-69-1, Molekula GmbH, Munich, Germany) was added to 0.02 mg membrane extracts prepared in 50 mM sodium phosphate buffer pH 4.6 to reach final concentrations of the antibiotic in the 0.00005–0.005 mg/mL range. Sample volume was 10 µL in all cases. Binding conditions were pH 4.6 for 10 min at 30°C, as described. Subsequently, 20 μM Boc-FL (Molecular Probes) was added and the sample incubated for 20 min at 30°C. Samples were finally processed by adding Laemmli buffer and boiled for 5 min. Proteins were resolved by SDS-PAGE in 8% (w/v) acrylamide gels. The gel was washed with 30% (v/v) methanol/10% (v/v) acetic acid, and fluorescence was detected on a Typhoon 8410 variable-mode imager (General Electric) with an excitation wavelength of 588 nm and a 520BP40 emission filter. Quantification of gel bands was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Bands with no signal changes in the presence of cefotiam were used as loading controls.
Microscopy analyses
Overnight bacterial cultures were diluted at OD600 ∼0.01 in LB pH 4.6 and grown in 96-well plates for 8 h in the presence of aztreonam (ref. 78110-38-0, Molekula GmbH, Munich, Germany) or cefotiam hydrochloride at varied concentrations. A volume of 120 μL of each culture was harvested (4300×g, 5 min, room temperature), washed in PBS pH 7.4 and fixed with 3% (w/v) paraformaldehyde. Images were acquired on an inverted Leica DMI 6000B microscope with an automated CTR/7000 HS controller (Leica Microsystems) and an Orca-R2 CCD camera (Hamamatsu Photonics).
Bacterial infection of fibroblasts
NRK-49F fibroblasts (ATCC CRL-1570) were propagated in DMEM containing 10% (v/v) FBS at 37°C in a 5% CO2 atmosphere as previously described. Fibroblasts were infected for 20 min at a multiplicity of infection of 10:1 with wild-type or mutant strains, all of them previously grown overnight in LB pH 4.6 without shaking. After this time, fibroblasts were washed three times with PBS pH 7.4, and fresh tissue culture medium containing 0.1 mg/mL gentamicin to kill remaining extracellular bacteria was added. At 2 hours post-infection (hpi), tissue culture medium was further replaced with fresh medium containing 0.01 mg/mL gentamicin and 0.001 mg/mL cefotiam hydrochloride. Cefuroxime at 0.02 mg/mL was also used as an alternative cephalosporin. Unlike cefotiam, cefuroxime has higher affinity for binding to PBP3 compared with PBP3SAL. At 2 and 24 hpi, fibroblasts were lysed in a 0.1% Triton X-100 solution and the number of viable intracellular bacteria was calculated by plating of 10-fold serial dilutions using PBS pH 7.4. Volumes plated were 100 μL onto LB pH 4.6 plates to calculate number of viable intracellular bacteria or 5 µL for the drop assay.
Statistical analyses
Data were analysed with GraphPad Prism, version 8.0, software (GraphPad Inc.). A t-test was used for data analysis. Significance was established at P values ≤0.05.
Results
Design of a screening method to identify drugs selectively inhibiting PBP3SAL versus PBP3
Unlike in E. coli, it is possible to generate S. Typhimurium mutants defective in PBP3 due to the presence of an alternative PBP, named PBP3SAL, which accomplishes cell division in acid pH. To select drugs targeting PBP3SAL with higher affinity than PBP3, we designed a screening method to search for compounds inducing filamentation, a manifestation of cell division blockage. The candidate compound was expected to cause filamentation in an S. Typhimurium ΔPBP3 mutant expressing only PBP3SAL for division but not in wild-type bacteria that express both PBP3 and PBP3SAL when growing in acidified (pH 4.6) LB medium. To detect differences in filamentation using high-throughput screening, we monitored the growth curve of wild-type and ΔPBP3 bacteria in the presence of aztreonam, a β-lactam that binds PBP3 with high affinity and blocks cell division at very low concentrations. The growth curves in the presence of a low dosage of aztreonam (0.001 mg/mL, equivalent to 2.3 µM) reached higher final optical density values (Figure 1a). This change was consistent with the presence of long filaments (Figure 1a). A dose–response assay for aztreonam confirmed the higher affinity for PBP3 compared with other PBPs. Thus, aztreonam caused S. Typhimurium filamentation starting at 0.0000078 mg/mL (0.018 µM) up to 0.05 mg/mL (115 µM), the highest concentration used (Figure S1, available as Supplementary data at JAC Online). No massive cell lysis was observed even at 115 µM of this antibiotic.
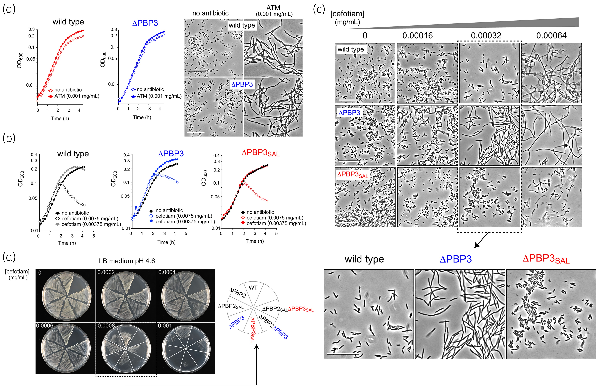
Figure 1
Identification of cefotiam in a screening designed to identify compounds that inhibit cell division by targeting PBP3SAL. (a) Higher final OD600 values detected in S. Typhimurium wild-type and ΔPBP3 isogenic strains incubated in the presence of 0.001 mg/mL (2.3 µM) aztreonam in LB medium pH 4.6. Microscope images were taken at 6 h after the onset of growth. Bar: 25 µm. (b) Effect on growth of S. Typhimurium wild-type, ΔPBP3 and ΔPBP3SAL isogenic strains incubated in LB medium pH 4.6 with the indicated concentrations of cefotiam hydrochloride. For these assays, the antibiotic was taken from the aliquot supplied at 10 mM in the Prestwick Chemical Library. (c) Effect of cefotiam on cell division visualized through the microscope in the indicated S. Typhimurium strains and antibiotic concentrations using LB medium pH 4.6. Note the differential effect when used at 0.00032 mg/mL (0.51 µM) causing blockage of cell division only in the strains producing PBP3SAL. The cefotiam used in these assays was acquired commercially with a purity of ≥98%. Bar: 25 µm. (d) Phenotype of the indicated isogenic S. Typhimurium single or double mutants lacking PBPs in response to commercially acquired cefotiam. Note the lack of growth at 0.0008 mg/mL (1.28 µM) cefotiam of the strains producing PBP3SAL as the only enzyme promoting cell division (ΔPBP3 genetic background). Assays were performed for a minimum of two independent biological replicates. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Based on these preliminary controls, we screened the Prestwick Chemical Library consisting of 1520 off-patent compounds. This library contains diverse anti-infective (antiviral, antiprotozoal, antifungal and antibacterial) compounds as well as drugs used in clinics to treat a number of pathologies, all supplied at 10 mM in DMSO. S. Typhimurium wild type and its isogenic ΔPBP3 mutant were exposed to these compounds at 1:100 dilution of the stock (100 µM). A few compounds increased final optical density values compared with control cultures not exposed to drug. These compounds were further discarded because no differential effect on wild-type and ΔPBP3 strains was observed. Compounds of this first group included β-lactam antibiotics like aztreonam, which validated our previous control assays (see Figures 1a and S1), cloxacillin, amoxicillin, piperacillin and cefazolin. A second group of β-lactams included in the Prestwick Library, comprising cefixime, cefotetan, ceforanide and cefotiam, caused lysis of wild-type and ΔPBP3 bacteria at 100 µM. Aiming to find different responses in these two strains, these β-lactams were subsequently tested at more diluted concentrations starting from 100 µM up to 3.125 µM. We also included an isogenic ΔPBP3SAL to discern whether the differences, if found, were related to the presence/absence of PBP3SAL.
Within this second group of β-lactams, cefotiam hydrochloride (MW = 598.56) was the only antibiotic causing a differential effect. In LB pH 4.6 medium, a condition in which S. Typhimurium wild type expresses PBP3 and PBP3SAL, cefotiam hydrochloride at 0.00375 mg/mL (6.25 µM) increased final optical density values in wild-type and ΔPBP3 strains, but not in the ΔPBP3SAL mutant (Figure 1b). We next acquired cefotiam hydrochloride powder at ≥98% purity from commercial sources. The response of bacteria exposed to cefotiam was then examined by microscopy in wild-type, ΔPBP3 and ΔPBP3SAL strains. Only those strains expressing PBP3SAL (wild-type and ΔPBP3) filamented in LB pH 4.6 medium containing 0.00032 mg/mL cefotiam hydrochloride whereas such an effect was not seen in the ΔPBP3SAL strain expressing only PBP3 (Figure 1c). The data obtained in liquid culture were confirmed in LB pH 4.6 agar plates containing distinct concentrations of cefotiam hydrochloride (Figure 1d). In the assays involving solid media we also tested isogenic single and double mutants lacking distinct PBPs involved in morphogenesis: PBP2, PBP3, PBP2SAL and PBP3SAL. Those strains having PBP3SAL as the single enzyme for division (ΔPBP3 genetic background) exhibited higher susceptibility to cefotiam (Figure 1d). Taken together, these observations suggested that the second-generation cephalosporin cefotiam could bind more efficiently to PBP3SAL than to PBP3.
Boc-FL bindings assays reveal higher affinity of cefotiam for PBP3SAL
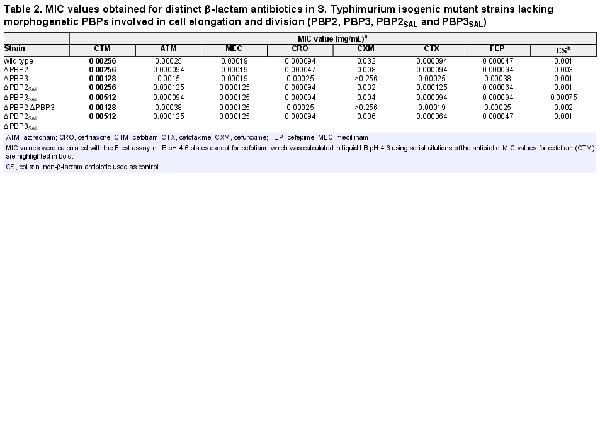
To further demonstrate the distinct behaviour of cefotiam for binding to PBP3 and PBP3SAL, a competition binding assay with the fluorescent derivative Boc-FL antibiotic was performed in membrane extracts obtained from ΔPBP3 and ΔPBP3SAL strains (Figure 2a). The assay revealed IC50 values for cefotiam of 0.0004 mg/mL in the case of PBP3SAL and 0.00375 mg/mL for PBP3 (Figure 2b). This result contrasted with our previous Boc-FL competition assays with other cephalosporins like cefuroxime, which exhibited higher affinity for PBP3. Cefotiam also showed avidity for binding to the high molecular weight bifunctional PBP1A and PBP1B, although with relatively high IC50 values of 0.004 and 0.0028 mg/mL for the ΔPBP3 and ΔPBP3SAL strains, respectively (Figure 2b). MIC values obtained for single and double mutants with deficiencies in PBP2, PBP3, PBP2SAL or PBP3SAL corroborated the higher susceptibility to cefotiam of those strains expressing PBP3SAL as the only PBP involved in cell division (Table 2). Conversely, the determination in S. Typhimurium by the Etest assay of MIC values for other cephalosporins known to have high affinity for PBP3, like aztreonam, ceftriaxone, cefuroxime, cefotaxime and cefepime, revealed an opposite phenotype compared with the response to cefotiam. Thus, the susceptibility to these other β-lactams increased notoriously in strains lacking PBP3SAL, i.e. having PBP3 as the only PBP for division (Table 2). The lack of PBP3SAL associated with increases in MIC values in the order of more than 8-fold (cefuroxime, cefepime), 6-fold (aztreonam) or 2.65-fold (ceftriaxone, cefotaxime) (Table 2). The MIC to mecillinam (amdinocillin), a β-lactam that binds specifically to PBP2 in E. coli, showed less than 2-fold difference in strains expressing either PBP3 or PBP3SAL (Table 2). The MIC to mecillinam was also very similar in strains expressing either PBP2 or PBP2SAL (Table 2). Unlike PBP3 and PBP3SAL, the pair PBP2/PBP2SAL might therefore bind mecillinam with similar affinity. Altogether, these data indicated that, unlike the other cephalosporins tested, cefotiam shows higher affinity for PBP3SAL compared with PBP3 and with no binding to other PBPs at concentrations in the 0.0001–0.001 mg/mL (0.16–1.6 µM) range.
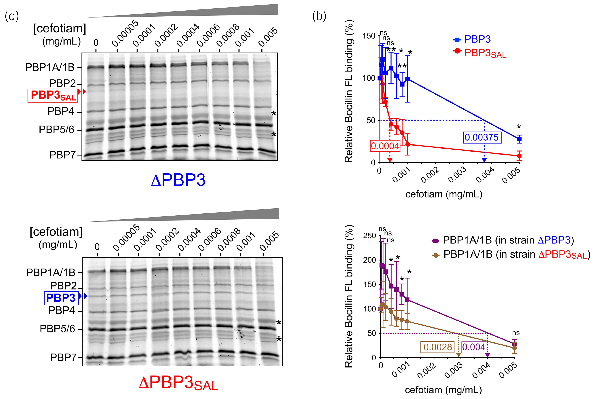
Figure 2
Cefotiam binds to PBP3SAL with higher affinity than to PBP3. (a) Representative BOCILLIN-FL competition binding assays performed in membrane extracts obtained from ΔPBP3 and ΔPBP3SAL isogenic mutants grown in LB medium pH 4.6. Binding conditions were also pH 4.6. Asterisks point to protein used in the densitometry analyses to adjust values for protein content. (b) Determination of the IC50 of cefotiam for competing BOCILLIN-FL binding to PBP1A/1B, PBP3 and PBP3SAL. For PBP1A/1B, data are shown separately corresponding to the data of ΔPBP3 and ΔPBP3SAL, respectively. Data are from three independent experiments and were analysed by Student’s t-test. *, P ≤ 0.05; **, P ≤ 0.005. ns, not significant. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Unlike cefuroxime, cefotiam is highly bactericidal against intracellular S. Typhimurium in a PBP3SAL-dependent manner
PBP3SAL is not produced by S. Typhimurium when growing extracellularly in neutral pH but its expression is up-regulated in acidic compartments of eukaryotic cells., Because cefotiam showed increased affinity for PBP3SAL, we reasoned that it could inhibit bacterial growth by blocking the activity of this PBP. To differentiate such predicted selectivity, we used the isogenic series of single and double S. Typhimurium mutants lacking PBP2, PBP3, PBP2SAL or PBP3SAL to infect the rat fibroblast NRK-49F cell line, in which we have extensively characterized mechanisms used by the pathogen to survive and persist intracellularly.,, Non-phagocytic cells consistent with fibroblasts are also targeted in vivo by S. Typhimurium in the intestinal lamina propria. The addition of 0.001 mg/mL (1.6 µM) cefotiam to the tissue culture medium at 2 hpi led to a drastic drop in the viability at 24 hpi of those strains dividing only with PBP3SAL (ΔPBP3 genetic background). This was demonstrated by drop assays performed on agar plates containing the antibiotic (Figure 3a) as well as by plating and cfu counting corresponding to viable intracellular bacteria at 2 hpi and 24 hpi (Figure 3b).
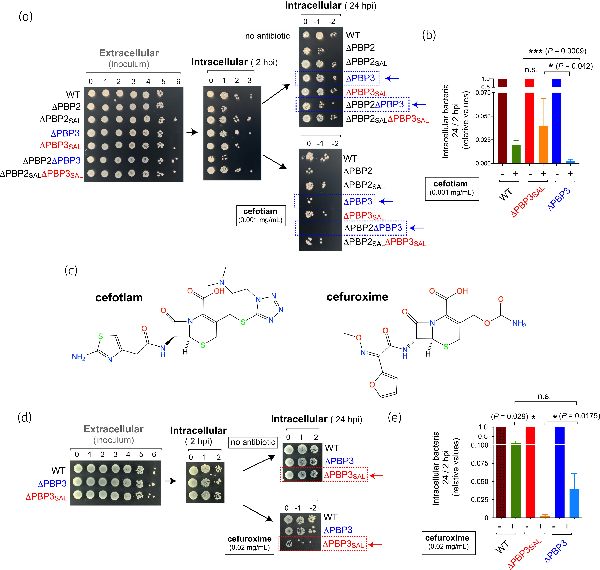
Figure 3
Cefotiam shows stronger viability inhibition in intracellular S. Typhimurium compared with other cephalosporins like cefuroxime. (a) Viability of the series of S. Typhimurium isogenic single and double mutants defective in PBPs inside NRK-49F rat fibroblasts at distinct post-infection times (2 hpi, 24 hpi) determined by the drop assay. Shown are serial dilutions of the overnight culture used for infection (inoculum) and the extracts obtained from the infected NRK-49F fibroblast culture. Cefotiam (0.001 mg/mL, 1.6 µM) was added to the tissue culture medium at 2 hpi. (b) Ratios of viable intracellular bacteria at 24 hpi versus 2 hpi determined for the indicated strains in the absence/presence of cefotiam. The values are shown relative to the samples not treated with the antibiotic and are the mean and SD of a total of the four independent biological replicates. Experimental mean of the 24 hpi/2 hpi ratios corresponding to four independent assays for samples without antibiotic were: 0.675 (wild type), 0.675 (ΔPBP3) and 0.738 (ΔPBP3SAL). (c) Structures of the cephalosporins cefotiam and cefuroxime, which show high affinity for PBP3SAL and PBP3, respectively. (d) Drop assay depicting the viability of S. Typhimurium wild-type, ΔPBP3 and ΔPBP3SAL strains inside NRK-49F fibroblasts at 2 hpi and 24 hpi. Shown are serial dilutions corresponding to inoculum (extracellular bacteria) and extracts containing intracellular bacteria. Cefuroxime (0.02 mg/mL, 47 µM) was added to the tissue culture medium at 2 hpi. (e) Ratios of viable intracellular bacteria at 24 hpi versus 2 hpi obtained for the indicated strains in the absence/presence of cefuroxime. The values are shown relative to the samples with no β-lactam added and are the mean and SD of four independent biological replicates. Data were analysed by Student’s t-test. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001. n.s., not significant. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
To further demonstrate the selective action of cefotiam against intracellular bacteria due to inhibition of PBP3SAL, we also tested cefuroxime (Figure 3c), a cephalosporin that has higher affinity for PBP3 than PBP3SAL. The bactericidal effect of cefuroxime in intracellular bacteria was the reverse to that of cefotiam, i.e. more pronounced for the ΔPBP3SAL mutant expressing PBP3 (Figure 3d,e). Overall, these data demonstrated that cefotiam is an antibiotic that reaches the intracellular compartment colonized by S. Typhimurium inside host cells and that it inhibits selectively PBP3SAL at the concentrations used.
Discussion
In enteric bacteria like E. coli and S. Typhimurium, PBP2 and PBP3 are morphogenetic peptidoglycan synthases directing the cell elongation and cell division phases, respectively., Due to these essential roles, PBP2 and PBP3 have been traditionally selected as targets for developing new drugs within the group of β-lactam antibiotics. Cephalosporins are widely used in clinics; however, some factors decrease their effectiveness. Besides the emergence of MDR and extensive-resistant isolates in S. enterica serovars with high incidence in humans such as Typhi, Typhimurium and Enteritidis, there are also reports of relapses in infections caused by antibiotic-susceptible isolates. This phenomenon is repeatedly documented in salmonellosis, with rates that can reach up to 15% of treated patients., Our previous work showed that S. Typhimurium mutants defective in PBP3SAL and, therefore, more susceptible to β-lactams, were less capable of causing relapse in a mouse typhoid model following ceftriaxone therapy. PBP3SAL is envisioned as a novel target of extreme importance in the biology of intracellular S. Typhimurium and consequently should be considered in future research directed to develop new effective antimicrobial drugs.
Unlike PBP3, PBP3SAL is expressed by the pathogen exclusively in acid environments, which precludes its analysis by the standard methods used in hospitals to determine antimicrobial susceptibility. In view of our findings, this practice, based on nutritional media at neutral pH like the Mueller–Hinton broth, should be revisited when analysing Salmonella isolates. Despite the unique feature of PBP3SAL of not binding antibiotics when expressed artificially at neutral pH and showing in acid pH reduced affinity for Boc-FL and other β-lactams like cefuroxime, this study provides evidence for the contrasting view of an antibiotic, cefotiam, with higher affinity to PBP3SAL than to PBP3. The encouraging data obtained in the in vitro infection model with cultured fibroblasts indicate that, when S. Typhimurium enters into a susceptible human or animal host, it should be possible to kill this pathogen ‘only’ in intracellular locations. The ∼10-fold difference in IC50 values for PBP3 and PBP3SAL obtained in the Boc-FL competition assays with cefotiam supports this possibility when using appropriate doses of the antibiotic. This innovative therapy should be theoretically innocuous to the endogenous microbiota given the absence of genes encoding PBP2SAL/PBP3SAL orthologues in the beneficial bacteria.
It is also worth recalling that cefotiam can be now be exploited to analyse computationally at the atomic level how it behaves as a ligand when entering the catalytic pocket of modelled PBP3 and PBP3SAL atomic structures. These analyses can provide insights into the different affinity found for this antibiotic to bind to these PBPs and facilitate the identification of new drugs acting with even higher selectivity. The structures of related cephalosporins that behave in an opposite manner to cefotiam regarding the bactericidal effect in intracellular bacteria, such as cefuroxime, are certainly of much value for the design of new antimicrobial drugs selectively controlling the intracellular infection.
Cefotiam was first described in 1979 as a second-generation cephalosporin more potent than others like cefazolin and being active against Gram-positive and Gram-negative bacteria, including Enterobacteriaceae genera such as Enterobacter spp., Klebsiella spp., E. coli, Salmonella spp. and Proteus. Some of these bacteria like E. coli and Proteus spp. do not have an alternative PBP3SAL-like homologue to PBP3. Furthermore, the antimicrobial susceptibility assays against these bacteria were performed in standard neutral pH (Mueller–Hinton medium), a condition in which at least in Salmonella spp., PBP3SAL is not expressed. These observations unequivocally demonstrate that cefotiam targets PBP3 at neutral pH, although probably at higher concentrations than those required to target PBP3SAL in acid pH. This difference regarding the effective inhibitory concentration in extra- and intracellular niches should be considered in future therapies based on cefotiam. Surprisingly, despite the optimal pharmacokinetics exhibited by cefotiam, to our knowledge no study has investigated the therapeutic potential of this cephalosporin to eradicate Salmonella infections. The data presented here, sustained with isogenic S. Typhimurium mutant strains lacking specific PBPs involved in cell elongation and division and alternative cephalosporins, clearly support the usage of cefotiam for treating salmonellosis.
Acknowledgements
We thank fruitful comments from members of García-del Portillo’s laboratory.
References
- 1. Castanheira S, García-Del Portillo F. Salmonella populations inside host cells. Front Cell Infect Microbiol2017; 7: 432. https://doi.org/10.3389/fcimb.2017.00432
- 2. Tulkens PM. Intracellular distribution and activity of antibiotics. Eur J Clin Microbiol Infect Dis1991; 10: 100–6. https://doi.org/10.1007/BF01964420
- 3. Peyrusson F, Nguyen TK, Buyck JM, et al In vitro models for the study of the intracellular activity of antibiotics. Methods Mol Biol2021; 2357: 239–51. https://doi.org/10.1007/978-1-0716-1621-5_16
- 4.
- 5. Ruby T, McLaughlin L, Gopinath S, et al Salmonella’s long-term relationship with its host. FEMS Microbiol Rev2012; 36: 600–15. https://doi.org/10.1111/j.1574-6976.2012.00332.x
- 6. Hill PWS, Moldoveanu AL, Sargen M, et al The vulnerable versatility of Salmonella antibiotic persisters during infection. Cell Host Microbe2021; 29: 1757–1773.e10. https://doi.org/10.1016/j.chom.2021.10.002
- 7. Ahmad KA, Khan LH, Roshan B, et al Factors associated with typhoid relapse in the era of multiple drug resistant strains. J Infect Dev Ctries2011; 5: 727–31. https://doi.org/10.3855/jidc.1192
- 8. Matono T, Kato Y, Morita M, et al Case series of imported enteric fever at a referral center in Tokyo, Japan: antibiotic susceptibility and risk factors for relapse. Am J Trop Med Hyg2016; 95: 19–25. https://doi.org/10.4269/ajtmh.15-0714
- 9. Rohs PDA, Bernhardt TG. Growth and division of the peptidoglycan matrix. Annu Rev Microbiol2021; 75: 315–36. https://doi.org/10.1146/annurev-micro-020518-120056
- 10.
- 11. Castanheira S, Cestero JJ, García-del Portillo F, et al Two distinct penicillin binding proteins promote cell division in different Salmonella lifestyles. Microb Cell2018; 5: 165–8. https://doi.org/10.15698/mic2018.03.622
- 12. Castanheira S, López-Escarpa D, Pucciarelli MG, et al An alternative penicillin-binding protein involved in Salmonella relapses following ceftriaxone therapy. EBioMedicine2020; 55: 102771. https://doi.org/10.1016/j.ebiom.2020.102771
- 13. Vivero A, Baños RC, Mariscotti JF, et al Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J Bacteriol2008; 190: 1152–6. https://doi.org/10.1128/JB.01206-07
- 14. Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature1981; 291: 238–9. https://doi.org/10.1038/291238a0
- 15. Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene1995; 158: 9–14. https://doi.org/10.1016/0378-1119(95)00193-A
- 16. Núñez-Hernández C, Tierrez A, Ortega AD, et al Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect Immun2013; 81: 154–65. https://doi.org/10.1128/IAI.01080-12
- 17. Georgopapadakou NH, Smith SA, Sykes RB. Mode of action of azthreonam. Antimicrob Agents Chemother1982; 21: 950–6. https://doi.org/10.1128/AAC.21.6.950
- 18. Neu HC. Penicillin-binding proteins and role of amdinocillin in causing bacterial cell death. Am J Med1983; 75: 9–20. https://doi.org/10.1016/0002-9343(83)90089-X
- 19. Núñez-Hernández C, Alonso A, Pucciarelli MG, et al Dormant intracellular Salmonella enterica serovar Typhimurium discriminates among Salmonella pathogenicity island 2 effectors to persist inside fibroblasts. Infect Immun2014; 82: 221–32. https://doi.org/10.1128/IAI.01304-13
- 20. López-Montero N, Ramos-Marquès E, Risco C, et al Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections. Autophagy2016; 12: 1886–901. https://doi.org/10.1080/15548627.2016.1208888
- 21. Egan AJF, Errington J, Vollmer W. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol2020; 18: 446–60. https://doi.org/10.1038/s41579-020-0366-3
- 22.
- 23. Castro-Vargas RE, Herrera-Sánchez MP, Rodríguez-Hernández R, et al Antibiotic resistance in Salmonella spp. isolated from poultry: a global overview. Vet World2020; 13: 2070–84. https://doi.org/10.14202/vetworld.2020.2070-2084
- 24. Pavelquesi SLS, de Oliveira Ferreira ACA, Rodrigues ARM, et al Presence of tetracycline and sulfonamide resistance genes in Salmonella spp. Literature review. Antibiotics (Basel)2021; 10: 1314.
- 25. Dragsted UB, Pedersen P. Relapse of multiresistant Salmonella typhi after combined therapy with ciprofloxacin and ceftriaxone. Clin Microbiol Infect2000; 6: 167–8. https://doi.org/10.1046/j.1469-0691.2000.00036.x
- 26. Parry CM, Hien TT, Dougan G, et al Typhoid fever. N Engl J Med2002; 347: 1770–82. https://doi.org/10.1056/NEJMra020201
- 27. Arjyal A, Basnyat B, Nhan HT, et al Gatifloxacin versus ceftriaxone for uncomplicated enteric fever in Nepal: an open-label, two-centre, randomised controlled trial. Lancet Infect Dis2016; 16: 535–45. https://doi.org/10.1016/S1473-3099(15)00530-7
- 28. Ogawa M, Hama M, Kosaki G, et al Comparison of cefotiam and cefazolin activity against Gram-negative bacilli. J Antimicrob Chemother1979; 5: 681–5. https://doi.org/10.1093/jac/5.6.681
- 29. Iwahi T, Tsuchiya K. Comparative activities of cefotiam and cefazolin against urinary tract infections with Proteus mirabilis in mice. Antimicrob Agents Chemother1980; 18: 257–63. https://doi.org/10.1128/AAC.18.2.257
- 30. Kan M, Wu L, Zhou X-W, et al Cefotiam treatment in children: evidence of subtherapeutic levels. Ther Drug Monit2020; 42: 733–6. https://doi.org/10.1097/FTD.0000000000000759
- 31. Brogard JM, Jehl F, Willemin B, et al Clinical pharmacokinetics of cefotiam. Clin Pharmacokinet1989; 17: 163–74. https://doi.org/10.2165/00003088-198917030-00003