Introduction
Infections caused by Gram-negative bacteria are increasingly difficult to treat due to the emergence and global spread of MDR or XDR strains., Of particular concern according to the WHO priority list is carbapenem-resistant Acinetobacter baumannii, against which novel antibiotics are urgently needed.
Rifabutin is a spiro-piperidyl rifamycin of the ansamycin antimicrobial drug class. Rifabutin (Mycobutin®) was approved by the FDA in 1992 as an oral formulation for the prevention of disseminated Mycobacterium avium complex disease in patients with advanced HIV infection. Rifabutin exerts its antibacterial activity by selective inhibition of the bacterial DNA-dependent RNA polymerase that catalyses RNA synthesis. Rifabutin was previously shown to have only minimal activity against Gram-negative bacteria when determined in standard nutrient-rich cationic-adjusted Mueller–Hinton broth (CA-MHB) because of the Gram-negative outer membrane that represents a permeability barrier. We found that rifabutin exerts potent antibacterial activity against A. baumannii when tested in the nutrient-limited Roswell Park Memorial Institute 1640 (RPMI) medium supplemented with FCS. This increased activity is mediated by cellular uptake of the antibiotic through the TonB-dependent siderophore receptor FhuE, which is up-regulated in RPMI supplemented with FCS but not in standard CA-MHB medium. Importantly, this novel in vitro activity correlated with excellent in vivo efficacy in murine systemic and lung infection models with A. baumannii. In this study we intended to determine the in vitro activity of rifabutin on a panel of recently isolated carbapenem-resistant A. baumannii (CRAB) isolates.
Materials and methods
Bacterial isolates
A. baumannii isolates (n = 293) used in this study were collected between 2017 and 2019 in various clinical laboratories from Europe (n = 144), the USA (n = 99) and Asia-West Pacific (n = 50) regions (JMI Laboratories, North Liberty, IA, USA). All the isolates were carbapenem resistant, and most of the isolates were from patients with bloodstream infection (n = 82), pneumonia (n = 174) or skin/soft tissue infection (n = 33) (Table S1, available as Supplementary data at JAC Online).
Susceptibility testing
Rifabutin MICs were determined using broth microdilution and agar dilution methods following the CLSI guidelines but using RPMI supplemented with 10% (v/v) FCS (IC-RPMI) as assay medium for broth microdilution and Mueller–Hinton agar (MHA) supplemented with 0.1 mM pyridoxal isonicotinoyl hydrazone (PIH) (IC-MHA) for agar dilution. MICs of the comparator antibiotics meropenem, ceftazidime, minocycline, colistin, tigecycline, tobramycin and cefiderocol were determined according to CLSI guidelines.
Gene expression level quantification
Quantitative RT-PCR (qRT-PCR) was performed to quantify gene expression levels as previously described. Expression was normalized to that of the housekeeping gene rpoD using the comparative ΔΔCT method. The list of primers used in this study can be found in Table S2.
Genetic engineering
All genetic engineering (deletion and allelic exchange) manipulations on A. baumannii clinical isolates were performed using a two-step recombination method previously described, as it allows manipulation of multiple clinical strains independent of their resistance profile. Briefly, DNA fragments corresponding to 700 bp upstream and downstream genomic regions of the genes to be deleted were amplified by PCR and cloned in the multiple cloning site of the knockout plasmid pVT77. For FhuE allelic exchange, the entire sequence from 700 bp upstream to 700 bp downstream of fhuEHUMC1 (including the fhuEHUMC1 sequence) was cloned in pVT77. The resulting plasmids were conjugated in A. baumannii isolates, and transconjugants were selected on LB agar plates containing 100 mg/L tellurite. The second recombination was selected on 200 mg/L 3′-azido-3′-deoxythymidine and the desired genetic modifications were confirmed by PCR and sequencing (Table S2).
Plasmid-mediated expression
The fhuE gene (AWC45_RS10145) from A. baumannii HUMC1 and LAC-4 strains and the arr-2 gene (ABAYE_RS17985) from the A. baumannii AYE strain were cloned into the Escherichia coli/A. baumannii shuttle plasmid pVT111, which contains the A. baumannii pWH1266 origin of replication, under the control of the IPTG-inducible promoter Ptrc-lacO. The FhuE V38P mutation (codon GTC to CCC) was introduced in FhuEHUMC1 by PCR site-directed mutagenesis. The resulting plasmids as well as the original pVT111 control plasmid were conjugated into A. baumannii ATCC 17978 and transconjugants were selected on LB agar plates containing kanamycin. The presence of the desired plasmids in A. baumannii was confirmed by PCR and sequencing (Table S2).
Genotyping of fhuE, rpoB and arr-2
A genomic DNA sequence including fhuE or the rifampicin resistance-determining regions (RRDRs) of rpoB was PCR amplified and the PCR products were sent for sequencing (Microsynth AG, Balgach, Switzerland). The full rpoB gene of strain BV667 was sequenced. The presence of arr-2 and fhuELAC-4 was assessed by PCR using primers specific for fhuELAC-4 that do not anneal on the fhuEHUMC1 allele (Table S2).
Determination of spontaneous resistance frequencies
The frequency of mutational resistance (FoR) to rifabutin was determined on MHA plates supplemented with 0.1 mM PIH. Cells from a 20 h culture in 50 mL of Mueller–Hinton medium were concentrated in 1 mL of PBS by centrifugation and 100 μL of 10-fold serial dilutions was inoculated on agar plates containing specified antibiotic concentrations. The plates were incubated at 35°C for 24 h and the FoR was calculated as the ratio between the number of colonies growing on plates with antibiotic and the total colony count of the inocula determined on non-selective agar plates.
Results
Iron chelation in standard Mueller–Hinton medium enables active rifabutin cellular uptake through FhuE overexpression
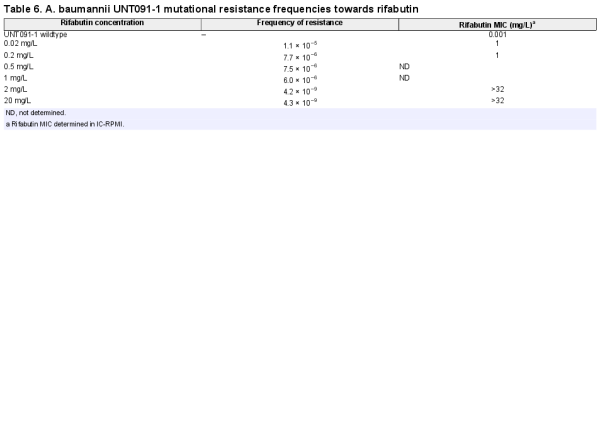
Potent rifabutin activity observed in RPMI supplemented with 10% FCS is enabled by FhuE overexpression in this specific nutrient- and iron-depleted medium. In contrast, the standard CA-MHB MIC testing medium is a complex and rich medium that does not promote FhuE expression (Figure 1a). Plasmid-mediated expression of FhuE in CA-MHB increased rifabutin activity to levels similar to those observed in RPMI supplemented with FCS, indicating that FhuE overexpression is a condition sufficient in CA-MHB to mediate rifabutin cellular uptake (Figure 2a, Figure S1). However, the activity of rifampicin and other known rifamycin antibiotics, including rifamycin CGP 4832, remained unchanged upon FhuE expression, indicating that FhuE is highly selective towards rifabutin transport (Figure S2). Moreover, the expression of a FhuE variant carrying a V38P mutation in the TonB box known to disrupt the interaction between FhuE and the energy transducer protein TonB, was not able to restore rifabutin susceptibility, indicating that the drug is actively transported via FhuE into A. baumannii (Figure 2a).,
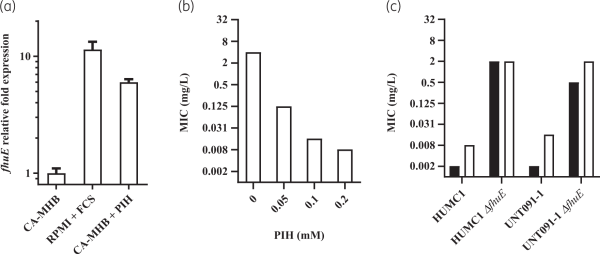
Figure 1
Quantification of fhuE expression level and rifabutin activity against A. baumannii. (a) A. baumannii HUMC1 fhuE transcript levels were determined in different media by quantitative real-time PCR and normalized to the fhuE expression of the strain grown in CA-MHB. CA-MHB and RPMI media were supplemented with 0.1 mM pyridoxal isonicotinoyl hydrazone (PIH) or 10% (v/v) FCS, respectively. (b) Rifabutin activity on A. baumannii HUMC1 determined in CA-MHB supplemented with PIH. (c) Rifabutin activity determined in RPMI supplemented with 10% (v/v) FCS (black) and MHA supplemented with 0.1 mM PIH (white) on WT A. baumannii strains and their ΔfhuE mutants.
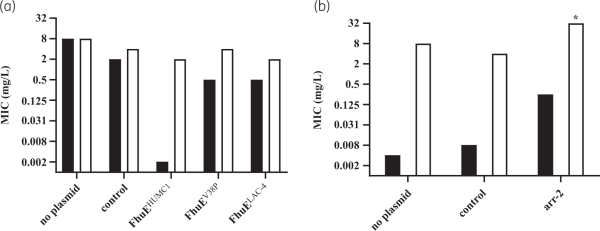
Figure 2
Activity of rifabutin (black bars) and rifampicin (white bars) upon plasmid-mediated expression of (a) different FhuE variants determined in CA-MHB medium and (b) Arr-2 determined in RPMI supplemented with 10% FCS. A. baumannii ATCC-17978 was used as host strain; gene expression from plasmids was induced with 1 mM IPTG, and a plasmid that did not encode fhuE or arr-2 was used as control. *MIC >32 mg/L.
Previous transcriptomic data have shown that reduction of available iron in rich medium using iron chelation led to fhuE overexpression in A. baumannii. We hypothesized that the iron present in CA-MHB must be chelated to achieve FhuE overexpression and potent rifabutin activity in order to develop a standard MIC testing medium that correlates with rifabutin in vivo efficacy. We used the iron chelator PIH to induce iron-chelated conditions in CA-MHB and demonstrated its ability to increase FhuE expression and rifabutin activity when added to CA-MHB (Figure 1a and b). PIH was non-toxic against A. baumannii, which was confirmed in a panel of diverse A. baumannii strains (Table S3). However, for some isolates with a very low rifabutin MIC, we noted the phenomenon of skipped wells with MICs determined in CA-MHB supplemented with PIH or with other iron chelators (apo-transferrin, 2,2-bipyridyl and deferiprone) (Figure S3). The characterization of clones from MIC with skipped wells demonstrated that this phenomenon results from mutations in fhuE leading to a disrupted rifabutin uptake (Table S4). Specific uptake disruption in vitro was previously reported for the antibiotic fosfomycin, for which agar dilution MIC is the only approved method allowing robust MIC determination; according to CLSI guidelines, broth microdilution should not be performed., Therefore, we determined rifabutin agar dilution MICs on MHA supplemented with 0.1 mM PIH and confirmed that PIH addition to MHA triggers a FhuE-dependent uptake of rifabutin (Figure 1c). The results obtained on MHA supplemented with 0.1 mM PIH are in accordance with the MICs observed in RPMI supplemented with 10% FCS, indicating that this iron-chelated MHA formulation (IC-MHA) allows a reliable determination of rifabutin MIC against A. baumannii. This standard agar dilution method may replace non-standard testing conditions using iron-chelated RPMI medium (IC-RPMI) to determine robust rifabutin MICs.
Rifabutin shows excellent activity on 293 CRAB isolates
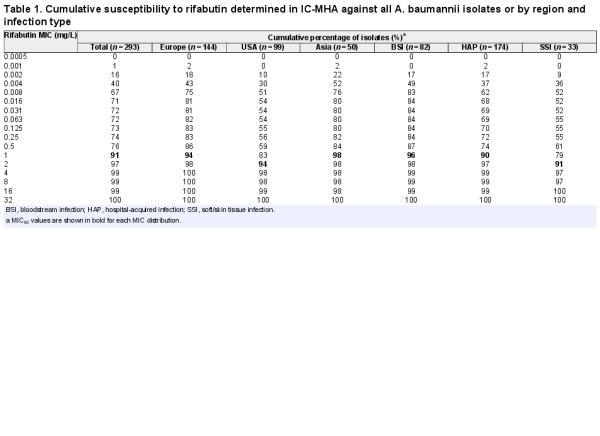
A panel of 293 CRAB clinical isolates collected between 2017 and 2019 from Europe (n = 144), the USA (n = 99) and Asia-West Pacific (n = 50) regions was used for rifabutin MIC determination. Rifabutin MICs were determined by broth microdilution and agar dilution methods in IC-RPMI and IC-MHA, respectively (Table S1). Rifabutin showed excellent activity in both testing methods, with an MIC50/MIC90 of 0.002/2 mg/L and 0.008/1 mg/L using IC-RPMI and IC-MHA, respectively (Figure 3a). There was little difference between testing methods in the overall susceptibility of the isolates. However, for the isolates with a very low MIC (≤0.008 mg/L), a one-dilution shift towards lower MIC in IC-RPMI was observed, which may reflect the nutrient-limited conditions of this medium (Figure 3a). In addition, rifabutin activity was consistent across regions and infection types, with only a one-dilution MIC90 shift for isolates from the USA or from soft/skin tissue infections (MIC90 of 2 mg/L) (Table 1).
Figure 3
Comparison of the efficacy of rifabutin against 293 carbapenem-resistant clinical isolates of A. baumannii in two different media. (a) Cumulative susceptibility and (b) MIC distribution of rifabutin determined in IC-RPMI and IC-MHA .
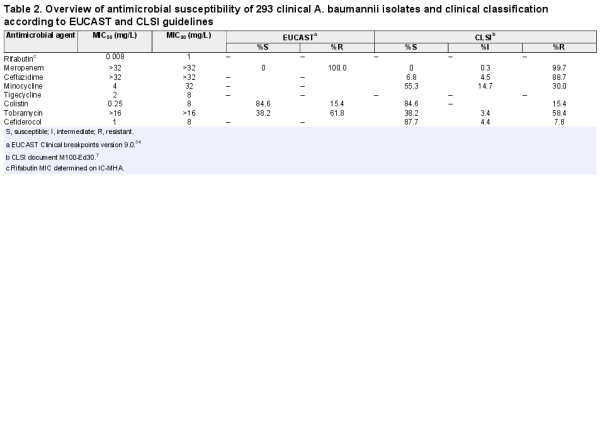
Rifabutin was superior to all comparator antibiotics tested, including colistin, tigecycline and cefiderocol (Figure 4, Table 2). Overall, rifabutin activity was not affected by resistance to specific antibiotics as the rifabutin MIC50/MIC90 remained stable on the different antibiotic-resistant subpopulations (Table 3). Importantly, there was no cross-resistance between rifabutin and cefiderocol despite both drugs employing bacterial iron transport mechanisms for efficient cellular uptake.
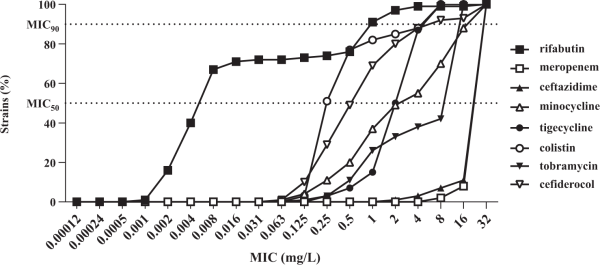
Figure 4
Cumulative susceptibility of rifabutin and comparator antibiotics against a panel of 293 carbapenem-resistant A. baumannii. Rifabutin MIC was determined in IC-MHA, whereas the MICs of meropenem, ceftazidime, minocycline, tigecycline, colistin, tobramycin and cefiderocol were determined according to CLSI guidelines.
Inactive rifabutin cellular uptake confers elevated rifabutin MIC in A. baumannii clinical isolates
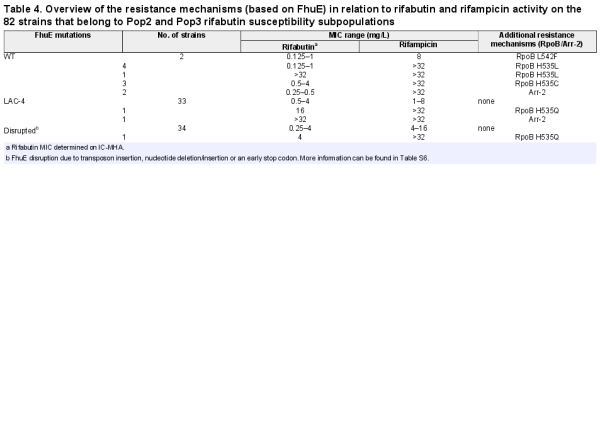
Further analysis of the rifabutin MICs revealed three A. baumannii subpopulations with distinct rifabutin susceptibilities, population 1 (Pop1, MICs <0.125 mg/L, 72%), population 2 (Pop2, MICs ≥0.125 to <16 mg/L, 27%) and population 3 (Pop3, MICs ≥16 mg/L, 1%) (Figure 3b, Table S5). The DNA sequence of the TonB-dependent siderophore receptor fhuE, responsible for rifabutin cellular uptake, was altered in 85% (n = 70) of the clinical isolates from Pop2 and Pop3 (n = 82) (Table 4, Table S6). Single mutation events inactivating FhuE were found in 50% (n = 35) of the strains with disrupted rifabutin uptake, including transposon insertions, nucleotide deletions/insertions and early stop codon mutations. The other 35 strains encoded a distinct FhuE variant (FhuELAC-4) with only 59% amino acid identity to the FhuE encoded in the reference strain HUMC1 (FhuEHUMC1).
Chromosomal replacement of fhuELAC-4 by the fhuEHUMC1 allele in the LAC-4 strain restored the potent activity of rifabutin (Figure 5a). In contrast, plasmid-mediated expression of FhuELAC-4 did not improve rifabutin activity in CA-MHB, confirming that FhuELAC-4 is unable to transport rifabutin (Figure 2a, Figure S1). To determine whether FhuELAC-4 was still active in transporting siderophores, we evaluated the ability of siderophores to restore the growth of the A. baumannii ATCC 17978 mutant deleted for endogenous acinetobactin biosynthesis and expressing FhuEHUMC1 or FhuELAC-4 in iron-limiting conditions, as previously described. Both FhuEHUMC1 and FhuELAC-4 variants restored growth in the presence of siderophores, indicating that despite their sequence divergence both FhuE variants can transport siderophores (Figure 6). The FhuELAC-4 variant may be derived from Acinetobacter pittii, which encodes a FhuE variant with 96% amino acid identity to FhuELAC-4, while the other pathogenic Acinetobacter species (namely A. nosocomialis, A. lactucae and A. seifertii) encode FhuE variants similar to FhuEHUMC1 (93%–97% amino acid identity). In addition, phylogenetic analysis revealed that each A. baumannii FhuE variant seems to be conserved in specific clonal lineages (Table S7). The FhuEHUMC1 variant is conserved in international clone (IC) 1 and IC2 strains, representing global clonal lineages, with IC2 being the most prevalent worldwide. In contrast, FhuELAC-4 is conserved in strains from IC5, which is a pan-American clonal lineage. This is in accordance with our strain panel where 80% of the strains encoding the FhuELAC-4 variant were isolated from the USA (Table S6).
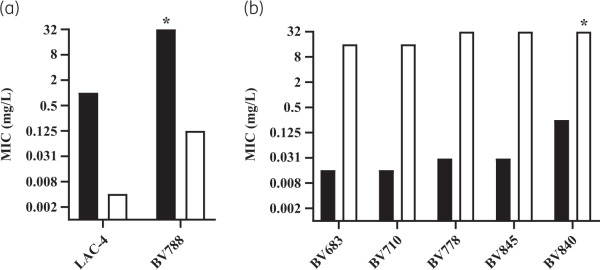
Figure 5
Rifabutin activity determined in IC-RPMI on A. baumannii strains with active or disrupted rifabutin uptake. (a) Rifabutin MIC on WT strains encoding the FhuELAC-4 variant (black bars) and their isogenic mutants where fhuELAC-4 was chromosomally exchanged for the fhuEHUMC1 allele (white bars). Strain BV788 carries an RpoB H535Q mutation while strain LAC-4 does not carry additional resistance mechanisms. *MIC >32 mg/L. (b) Rifabutin MIC on WT strains encoding the FhuEHUMC1 variant (black bars) and their ΔfhuE isogenic mutants (white bars). Strains BV683, BV710, BV778 and BV845 carry RpoB mutations I581M, S583L, H535Q and N527D, respectively, and strain BV840 encodes Arr-2 inactivation enzyme. *MIC >32 mg/L.
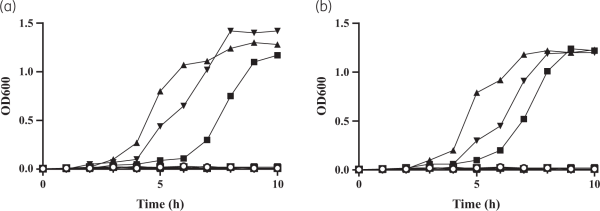
Figure 6
Growth complementation of the A. baumannii mutant strain depleted for endogenous siderophore production to determine the ability of FhuEHUMC1 and FhuELAC-4 variants to transport siderophores. A. baumannii ATCC 19798 ΔbasDΔfhuE double mutant strains complemented with (a) FhuEHUMC1-expressing plasmid or (b) FhuELAC-4-expressing plasmid were grown in LB medium containing 225 μM 2,2-bipyridyl supplemented with 20 μM desferricoprogen (square), rhodotorulic acid (triangle), desferrioxamine B (inverted triangle) or no siderophores (circle), as previously described.A. baumannii ATCC 19798 ΔbasDΔfhuE double mutant strains complemented with an empty plasmid were used as controls (open symbols).
Rifabutin overcomes RpoB mutations and Arr-2 expression conferring rifampicin resistance in A. baumannii clinical isolates
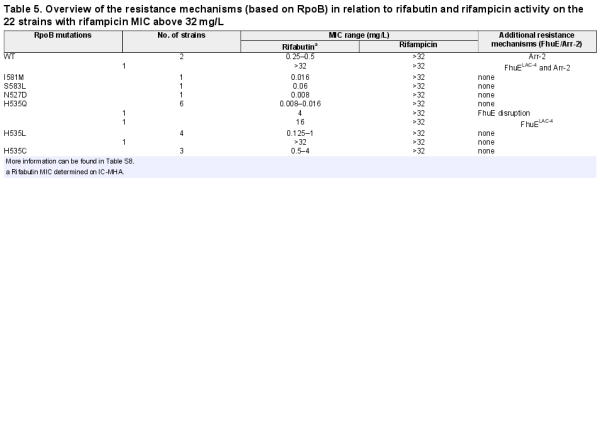
Mutations in the DNA-dependent RNA polymerase β-subunit (RpoB) and expression of the arr-2 gene coding for rifampicin ADP-ribosyltransferase are mechanisms that confer resistance to rifamycin antibiotics, and those resistance mechanisms have previously been identified in A. baumannii. Mutations in RpoB RRDR cluster I and II were identified in 86% (n = 19) of the strains with an elevated rifampicin MIC (MIC >32 mg/L, n = 22) while the other 14% (n = 3) of the strains with an elevated rifampicin MIC encoded the arr-2 gene (Table 5, Table S8).
The most frequent RpoB mutation, found in 16 isolates (73%), was the substitution of a histidine at position 535 by a glutamine (n = 8), leucine (n = 5) or cystine (n = 3). Among the isolates with RpoB mutations, nine strains belonged to Pop1 rifabutin susceptibility (MICs <0.125 mg/L), eight strains belonged to Pop2 (MICs ≥0.125 to <16 mg/L) and only two strains belonged to Pop3 (MICs ≥16 mg/L), suggesting that most RpoB mutations present in A. baumannii clinical isolates do not abolish rifabutin activity, in contrast to rifampicin loss of activity. Rifabutin uptake deletion in the Pop1 strains carrying RpoB mutations converted them to Pop3 strains, suggesting that potent rifabutin activity on RpoB mutants is due to FhuE-mediated active transport of rifabutin (Figure 5b). Interestingly, the two Pop3 strains (BV667 and BV788) carried RpoB mutations identical to other Pop1 and Pop2 strains, suggesting that an additional mechanism must be involved in the elevated rifabutin MIC of these two strains (Table 5). The strain BV788 encoded a FhuELAC-4 variant in addition to the RpoB H535Q mutation, and chromosomal replacement of fhuELAC-4 by fhuEHUMC1 in this strain confirmed that rifabutin overcomes resistance conferred by this RpoB mutation (Figure 5a). The second Pop3 strain (BV667) carried an RpoB H535L mutation but did not encode additional arr-2 or fhuE disruption, indicating that another rifabutin resistance mechanism may exist. The entire rpoB gene of BV667 was sequenced to rule out additional mutations outside RRDR clusters, and the induction of fhuE expression upon iron chelation was confirmed (Figure S4).
Among the arr-2-expressing isolates, two strains belonged to rifabutin susceptibility Pop2 and only one strain belonged to Pop3 (Table 5). The Pop3 strain encoded a FhuELAC-4 variant in addition to the arr-2 gene. Deletion of fhuE in a strain carrying arr-2 (BV840) as its only resistance mechanism, as well as plasmid-mediated arr-2 expression, demonstrated that the arr-2 resistance mechanism alone does not abolish rifabutin activity, in contrast to loss of rifampicin activity (Figures 2b and 5b).
The frequency of mutational resistance of clinical A. baumannii strains to rifabutin is dose dependent, reaching 10−9 at drug concentrations at or above 2 mg/L
The mutational FoR to rifabutin determined on the Pop1 strain UNT091-1 ranged from 10−5 to 10−6 with rifabutin concentrations up to 1 mg/L and decreased to 10−9 at rifabutin concentrations at or above 2 mg/L (Table 6). No difference was observed between rifabutin FoR determined on IC-MHA and IC-RPMI agar plates. This dose-dependent FoR suggests that a multistep mutational process is required to acquire full rifabutin resistance (MIC >32 mg/L), which is in accordance with the identification of at least two independent resistance mechanisms in Pop3 clinical isolates (Tables 4 and 5). In addition, the median FoR determined at 4-fold MIC on 10 Pop2 strains was 6.7×10−9, indicating that intermediate resistance levels do not affect the FoR to rifabutin (Table S9). Finally, FoRs to rifabutin were 1 log lower than FoRs to meropenem and minocycline and in the same range as the FoR to tobramycin (Table S10). Our data suggest that rifabutin resistance development in A. baumannii is low at or above 2 mg/L, and similar or better than resistance development to standard of care antibiotics.
Discussion
Rifabutin demonstrated potent in vitro activity on clinical CRAB isolates collected between 2017 and 2019 from three different continents. Rifabutin was equally active across geographical regions and infection types, including bloodstream infection and hospital-acquired pneumonia, with an MIC90 of 1–2 mg/L. The clonal nature of A. baumannii may lead to a considerable number of identical strains derived from clinical outbreaks. However, the tested isolates were collected from 72 different clinical sites within 22 different countries to reduce a potential bias of the study. In terms of MIC90, rifabutin was superior to all tested comparator antibiotics including colistin and cefiderocol. Notably, rifabutin maintained potency against A. baumannii subpopulations resistant to different classes of antibiotics, such as strains resistant to colistin, minocycline, tobramycin and cefiderocol. The absence of cross-resistance between rifabutin and the siderophore–drug conjugate cefiderocol suggests that these two drugs use different iron uptake systems for cellular influx. Cefiderocol transport systems have been identified in E. coli (CirA and Fiu) and in Pseudomonas aeruginosa (PiuA) but not in A. baumannii. Nevertheless, cefiderocol resistance in A. baumannii is mainly due to the expression of specific β-lactamases, justifying the absence of cross-resistance to rifabutin.
Rifabutin MICs determined in IC-MHA medium were consistent with MICs determined in IC-RPMI, in which the potent activity of rifabutin was originally observed. The rifabutin broth microdilution MIC method using IC-CA-MHB (CA-MHB supplemented with 0.1 mM PIH) medium sometimes led to skipped wells due to rifabutin uptake mutations. Interestingly, this phenomenon was observed only with highly susceptible strains (Pop1) and at rifabutin concentrations below 2 mg/L, which is in line with the low FoR observed at or above 2 mg/L. Skipped wells in the broth microdilution MIC method translates in agar dilution MIC tests to single colony growth, which, according to the CLSI, should be disregarded when reading agar dilution MIC endpoints. Therefore, the agar dilution MIC method using IC-MHA medium allows robust determination of rifabutin activity on all A. baumannii strains while the broth microdilution MIC method using IC-CA-MHB medium allows robust determination of rifabutin MICs only in Pop2 and Pop3 strains (MIC ≥0.125 mg/L).
The trimodal MIC distribution and the dose-dependent FoR suggested that resistance to rifabutin is a multistep process. This was confirmed with the characterization of rifabutin resistance mechanisms in the Pop2 and Pop3 strains. All the strains with rifabutin MIC at or above 0.125 mg/L had disrupted rifabutin uptake (FhuE), a mutated rifabutin target (RpoB) or a rifabutin inactivation enzyme (Arr-2). Notably, we demonstrated that loss of rifabutin activity (MIC >32 mg/L) was conferred by a combination of at least two independent resistance mechanisms in clinical strains. In contrast, RpoB mutations or the presence of the Arr-2 inactivation enzyme led to loss of rifampicin activity (MIC >32 mg/L). It has been described that rifabutin retains activity in the case of specific RpoB mutations that confer rifampicin resistance in Mycobacterium tuberculosis. However, disruption of rifabutin uptake in A. baumannii Pop1 strains carrying RpoB mutations decreases rifabutin activity to the same extent as rifampicin. Moreover, Arr-2 expression has been shown to confer equal resistance to rifampicin and rifabutin. Altogether these data suggest that rifabutin overcomes RpoB mutation and Arr-2 expression resistance mechanisms because it is actively transported into A. baumannii. We propose a rifabutin mechanism of action in which, as in contrast to rifampicin passive diffusion, FhuE-mediated rifabutin active transport creates a high concentration of drug at the site of action, enabling activity against target mutation and enzymatic inactivation resistance mechanisms (Figure 7). The identification of one Pop3 strain only carrying the RpoB H535L mutation, which is also present in several Pop2 strains, suggests that rifabutin resistance mechanisms not identified in this study may exist in clinical isolates.
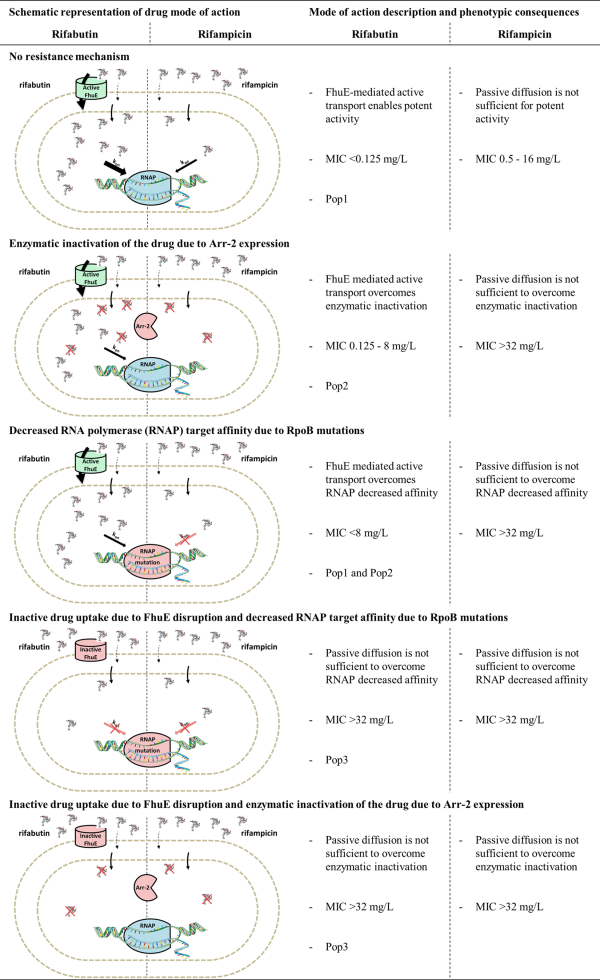
Figure 7
Model for rifabutin and rifampicin mode of action and resistance mechanisms in A. baumannii. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
TonB-dependent transporter (TBDT)-mediated uptake of the rifamycin derivative CGP 4832 has previously been described. Rifamycin CGP 4832 is actively transported across the outer membrane of E. coli by the siderophore receptor FhuA, but we demonstrated that it is not transported by A. baumannii FhuE. Conversely, rifabutin seems not to be transported by any TBDT from E. coli, Klebsiella pneumoniae and P. aeruginosa, as rifabutin does not show potent activity against those Gram-negative species. In addition, none of the commercially available rifamycin antibiotics was transported by A. baumannii FhuE. These results are in line with TBDT high substrate specificity, as TBDT-mediated transport requires specific substrate binding to activate allosteric conformational transition of the transporter leading to the recruitment of the TonB energy transducing machinery through the so-called TonB box of the TBDT. It has been demonstrated that rifamycin CGP 4832 is transported in a TonB-dependent manner, and a crystallographic study suggested that the binding of CGP 4832 to FhuA induces an allosteric transition allowing physical interaction between TonB and the FhuA TonB box. In the same manner, we demonstrated that rifabutin is transported by FhuE in a TonB-dependent manner, suggesting that rifabutin binding to FhuE is required to induce active transport through the A. baumannii outer membrane.
Our in vitro data on rifabutin activity and mutational resistance frequency suggest that rifabutin concentrations must be at or above 2 mg/L to be effective against A. baumannii infections and to avoid rapid resistance development. Rifabutin is available on the market as an oral drug with a maximum recommended dose of 300 mg/day. However, pharmacokinetic studies have shown that maximum concentrations of rifabutin remained below 1 mg/L with a daily dose up to 600 mg. Therefore, use of oral rifabutin in the treatment of A. baumannii infections will have only limited efficacy and would certainly lead to rapid development of resistance. In an attempt to overcome these limitations, BioVersys AG has developed an intravenous formulation of rifabutin (BV100) for the treatment of A. baumannii infections that have limited treatment options, including hospital- and ventilator-associated pneumonia and bloodstream infections, for patient populations where an intravenous formulation is required.
Acknowledgements
JMI Laboratories provided the panel of 293 carbapenem-resistant A. baumannii strains used in this study.
Funding
This study was supported by internal funding. V.L. was supported by the European Commission via the International Training Network Train2Trarget (721484).
Transparency declarations
All authors are employees and own shares or stock options of BioVersys AG.
References
- 1. Boucher HW, Talbot GH, Bradley JS et al Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 1–12.
- 2. Vasoo S, Barreto JN, Tosh PK. Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin Proc 2015; 90: 395–403.
- 3. Tacconelli E, Carrara E, Savoldi A et al Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18: 318–27.
- 4. Crabol Y, Catherinot E, Veziris N et al Rifabutin: where do we stand in 2016? J Antimicrob Chemother 2016; 71: 1759–71.
- 5. Vaara M. Comparative activity of rifabutin and rifampicin against Gram-negative bacteria that have damaged or defective outer membranes. J Antimicrob Chemother 1993; 31: 799–801.
- 6.
- 7. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—30th Edition: M100-Ed30. 2020.
- 8. Trebosc V, Gartenmann S, Royet K et al A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob Agents Chemother 2016; 60: 7263–71.
- 9. Funahashi T, Tanabe T, Mihara K et al Identification and characterization of an outer membrane receptor gene in Acinetobacter baumannii required for utilization of desferricoprogen, rhodotorulic acid, and desferrioxamine B as xenosiderophores. Biol Pharm Bull 2012; 35: 753–60.
- 10. Cadieux N, Bradbeer C, Kadner RJ. Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J Bacteriol 2000; 182: 5954–61.
- 11. Eijkelkamp BA, Hassan KA, Paulsen IT et al Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 2011; 12: 126.
- 12. Ballestero-Téllez M, Docobo-Pérez F, Rodríguez-Martínez JM et al Role of inoculum and mutant frequency on fosfomycin MIC discrepancies by agar dilution and broth microdilution methods in Enterobacteriaceae. Clin Microbiol Infect 2017; 23: 325–31.
- 13. Ito A, Nishikawa T, Matsumoto S et al Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2016; 60: 7396–401.
- 14. Higgins PG, Dammhayn C, Hackel M et al Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 2010; 65: 233–8.
- 15. Giannouli M, Di Popolo A, Durante-Mangoni E et al Molecular epidemiology and mechanisms of rifampicin resistance in Acinetobacter baumannii isolates from Italy. Int J Antimicrob Agents 2012; 39: 58–63.
- 16. Baysarowich J, Koteva K, Hughes DW et al Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc Natl Acad Sci USA 2008; 105: 4886–91.
- 17. Fournier P-E, Vallenet D, Barbe V et al Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2006; 2: e7.
- 18. Houang ETS, Chu Y-W, Lo W-S et al Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-β-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob Agents Chemother 2003; 47: 1382–90.
- 19. Ito A, Sato T, Ota M et al In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob Agents Chemother 2018; 62: e01454–17.
- 20. Yamano Y. In vitro activity of cefiderocol against a broad range of clinically important Gram-negative bacteria. Clin Infect Dis 2019; 69: S544–51.
- 21. CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition: M07-A10. 2015.
- 22. Yang B, Koga H, Ohno H et al Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J Antimicrob Chemother 1998; 42: 621–8.
- 23. Williams DL, Spring L, Collins L et al Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 1998; 42: 1853–7.
- 24. Cavusoglu C, Karaca-Derici Y, Bilgic A. In-vitro activity of rifabutin against rifampicin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Clin Microbiol Infect 2004; 10: 662–5.
- 25. Farhat MR, Sixsmith J, Calderon R et al Rifampicin and rifabutin resistance in 1003 Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother 2019; 74: 1477–83.
- 26. Whitfield MG, Warren RM, Mathys V et al The potential use of rifabutin for treatment of patients diagnosed with rifampicin-resistant tuberculosis. J Antimicrob Chemother 2018; 73: 2667–74.
- 27. Jing W, Pang Y, Zong Z et al Rifabutin resistance associated with double mutations in rpoB gene in Mycobacterium tuberculosis isolates. Front Microbiol 2017; 8: 1768.
- 28. Rukasha I, Said HM, Omar SV et al Correlation of rpoB mutations with minimal inhibitory concentration of rifampin and rifabutin in Mycobacterium tuberculosis in an HIV/AIDS endemic setting, South Africa. Front Microbiol 2016; 7: 1947.
- 29. Wehrli W, Zimmermann W, Kump W et al CGP 4832, a new semisynthetic rifamycin derivative highly active against some Gram-negative bacteria. J Antibiot 1987; 40: 1733–9.
- 30. Pugsley AP, Zimmerman W, Wehrli W. Highly efficient uptake of a rifamycin derivative via the FhuA–TonB-dependent uptake route in Escherichia coli. J Gen Microbiol 1987; 133: 3505–11.
- 31. Ferguson AD, Ködding J, Walker G et al Active transport of an antibiotic rifamycin derivative by the outer-membrane protein FhuA. Structure 2001; 9: 707–16.
- 32. Noinaj N, Guillier M, Barnard TJ et al TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 2010; 64: 43–60.
- 33. Blaschke TF, Skinner MH. The clinical pharmacokinetics of rifabutin. Clin Infect Dis 1996; 22 Suppl 1: S15–21; discussion S21–2.
- 34. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0, 2020.