Introduction
Species of the genus Echinostoma Rudolphi, 1809 are digenetic trematodes found, in the adult stage, in the intestine of birds and mammals, including humans (Fried and Graczyk, ; Toledo et al., ; Chai et al., ). More than 120 nominal species have been described worldwide making it the most diverse genus in the Echinostomatidae (Kostadinova and Gibson, ), but the pronounced morphological uniformity among species, along with other historical issues, has led to a complex taxonomic scenario. This is particularly true for the ‘revolutum’ group, formed by species of Echinostoma possessing 37 collar spines. About 60 nominal species were described worldwide (Chai et al., ), but the validity of most of them still requires confirmation, given that this group has been subjected to intense debate and constant taxonomic rearrangements over time (Georgieva et al., , ; Faltýnková et al., ).
In the last decades, the use of molecular tools has revealed the existence of numerous independent evolutionary lineages of Echinostoma, usually referred to as cryptic species, but also has helped to unravel the correct identity of previously known taxa, some of them described as new (Detwiler et al., ; Georgieva et al., , , ; Faltýnková et al., ; Izrailskaia et al., ; Valadão et al., ). These recent advances have contributed to a better knowledge of the species composition and the distribution of these parasites and have generated evidence on their biogeography, evolution and radiation. However, such integrative taxonomic studies involving Echinostoma spp. are disproportionately carried out in Asia, Europe and North America (Detwiler et al., ; Georgieva et al., ; Chai et al., ). In fact, in South America, molecular studies involving species of Echinostoma are scarce (Maldonado et al., ; Valadão et al., ) and do not reflect the diversity of species known in this subcontinent (more than 30 species, 12 bearing 37 collar spines) (Travassos et al., ; Fernandes et al., ; Lunaschi et al., ; Valadão et al., ).
Among the species belonging to the ‘revolutum’ group is Echinostoma paraensei Lie and Basch, 1967, described based on adult parasites recovered in the course of experimental studies initiated from naturally infected Biomphalaria glabrata (Say, 1818) collected in Brazil. The main differential trait of the species is the relatively small size of the dorsal collar spines (Lie and Basch, ). Some decades later, the semi-aquatic rodent, Nectomys squamipes (Brants, 1827), was identified as a natural definitive host in Brazil (Maldonado et al., ). The life cycle of the species is maintained under laboratory conditions in the USA and Brazil (Toledo and Fried, ; Fried and Peoples, ), which made it possible to carry out several biological studies involving E. paraensei. These studies contributed to the knowledge of several aspects related to this species, including (i) parasite behaviour (e.g. Loker et al., ; Nollen, ; Maldonado et al., ), (ii) parasite interaction (in both intramolluscan and vertebrate phases) with Schistosoma mansoni Sambon, 1907 and/or other echinostomes (e.g. Sullivan and Richards, ; Combes, ; Maldonado et al., ; Garcia et al., ; Hanington et al., ), (iii) ultrastructural morphology of different developmental stages (e.g. Fujino et al., ; Maldonado et al., ; Pinheiro et al., ; Souza et al., ), (iv) biochemical and pathological changes associated with the infection in molluscs and rodents (e.g. Adema et al., ; Toledo, ; Garcia et al., , ; Souza et al., ), (v) parasite protein composition (proteomics) (e.g. Loker et al., ; Marcilla, ) and (vi) sequencing of expressed sequence tags (Adema et al., ; Nowak and Loker, ).
Despite this tremendous scientific knowledge accumulated over decades about E. paraensei, studies on the molecular characterization of the species are scarce. Considering genetic markers, commonly used in taxonomic studies of trematodes, few sequences generated for E. paraensei are available in the public genetic sequence databases. They correspond to the complete mitochondrial genome of the isolate maintained under laboratory conditions in the USA (KT008005 – Adema et al., direct submission in GenBank®, unpublished), some internal transcribed spacer (ITS) sequences generated for the Brazilian isolate from N. squamipes (Maldonado et al., ) and nad1 sequences obtained for larval stages found in Australia and for the isolate originated from Brazil and maintained in the USA (Morgan and Blair, ).
In the present study, we provide data obtained during experimental, morphological and molecular studies of a 37-collar-spined echinostome found in Stenophysa marmorata (Guilding, 1828) from Brazil. Despite the morphological data of the developmental stages and sequences of nuclear genetic markers (28S and ITS) suggested that the parasite is indistinguishable from E. paraensei, mitochondrial data (nad1 gene) pointed to a genetically distinct lineage, herein described as a new species of Echinostoma.
Materials and methods
Sampling and experimental infection
The isolate of the parasite evaluated in this study was obtained from a specimen of S. marmorata collected in the municipality of Alvorada de Minas, Minas Gerais, Brazil, in February 2019 during a malacological survey focused on the epidemiology of schistosomiasis. Details of malacological techniques and preliminary evaluation of infection with larval trematodes are as described in Coelho et al. (, ). The infected snail was sent to the laboratory for further parasite identification. A new photostimulation test was carried out for the recovery of cercariae. The infected mollusc died on the following day and was dissected for retrieval of rediae and metacercariae. This fact precludes us from providing detailed illustrations of larval stages.
For the recovery of adult parasites, part of the collected metacercariae were used to infect a laboratory-reared jird, Meriones unguiculatus (Milne-Edwards, 1867). The rodent was orally inoculated with approximately 60 metacercariae. The infected animal was submitted to an immunosuppression protocol through subcutaneous inoculation of dexamethasone disodium phosphate (Decadron, Ache, Brazil), 25 mg kg−1, daily until the end of the experiment. The success of infection was verified by fecal examination through the spontaneous sedimentation method (Lutz, ) from 7 days post-infection (dpi) onwards. The animal was kept in the laboratory with water and food provided ad libitum until the euthanasia carried out at 15 dpi. The intestine was transferred to Petri dishes containing physiological saline (0.85% NaCl) and examined under a stereomicroscope for the presence of adult or juvenile parasites.
Morphological study
Samples of the emerged cercariae were stained with vital stains (0.05% neutral red or Nile blue sulphate) and mounted in temporary preparations for examination under a light microscope. Larvae were also killed in hot water and fixed in 10% formalin for morphometric analysis. Rediae and metacercariae were studied live in temporary preparations and then also fixed in 10% formalin. Adult parasites collected from the experimentally infected jird were individually compressed between glass slides and fixed in 10% formalin. Later, they were stained with alum acetocarmine, dehydrated in an increasing series of ethanol, clarified in beechwood creosote and mounted on permanent slides with Canada balsam. For the study of the cephalic collar, some specimens had their anterior extremity sectioned with the aid of a scalpel blade, transferred to a glass slide and clarified with Amman's lactophenol. The preparations were examined under the microscope for determination of the number, size, shape and arrangement of collar spines, according to Fried et al. (). Whole adult parasites were photographed using a digital camera Samsung® ES70 (Samsung, Brazil), coupled to a stereomicroscope. Details of structures of adults and the larval stages were photographed with a Leica ICC50 HD digital camera, coupled to a Leica DM500 microscope. Captured images were analysed with Leica Application Suite software (LAZ EZ), version 2.0 (Leica Microsystems, Germany). The drawings were made with the aid of a camera lucida connected to an Olympus BH-2 microscope (Olympus, Japan). The morphometric analyses were carried out using a micrometre eyepiece. The abbreviations used in the morphological descriptions are in accordance with the most recent descriptions of Echinostoma (Kostadinova, , Faltýnková et al., ; Georgieva et al., ). In the case of dorsal spines (DLSL, length of the dorsolateral spines; DMSL, length of the most dorsal spines), we used the terminology presented by Maldonado et al. (). Data obtained were compared with descriptions available to Echinostoma spp. reported in South America (Lie and Basch, ; Travassos et al., ; Maldonado et al., , ; Fernandes et al., ).
The type material is deposited in the Helminthological Collection of the Institute Oswaldo Cruz (CHIOC) and Collection of Trematodes of the Federal University of Minas Gerais (UFMG-TRE).
Molecular study
An adult specimen fixed in 95% ethanol was used for DNA extraction using the QIAamp® DNA Mini kit (Qiagen, USA), according to manufactures' instructions. The concentration and quality of extracted DNA was evaluated using the microvolume spectrophotometer NanoDrop® Lite (Thermo Fisher Scientific, Rockford, IL, USA). Partial regions of 28S rRNA gene (primers Dig-12/1500R; Tkach et al., ), ITS1-5.8S-ITS2 regions (primers BD1/BD2; Luton et al., ), cox1 gene (JB3/CO1-R trema; Miura et al., ) and nad1 gene (primers NDJ11/NDJ2a; Kostadinova et al., ) were amplified by PCR. The amplification reactions were carried out in a final volume of 25 μL, including 12.5 μ,L of Platinum Hot Start Master Mix 2 × (Invitrogen, Thermo Fisher Scientific Baltics, Vilnius, Lithuania), 1.25 pmol of each primer, about 50 ng of template DNA and sterile ddH2O. PCR products were electrophoresed on a 1.5% agarose gel, and the band of the expected size was purified with 20% polyethylene glycol 8000 (Promega, Madison, WI, USA). Purified DNA was sequenced in both directions by capillary electrophoresis in an ABI3730 sequencer using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). The chromatograms obtained were assembled using ChromasPro v.2.0.1 software (Technelysium Pty Ltd, South Brisbane, Queensland, Australia) and the generated contiguous sequences used in the subsequent analyses.
The newly generated sequences were first subjected to a search in the Basic Local Alignment Search Tool – BLAST (Altschul et al., ) (https://blast.ncbi.nlm.nih.gov) in GenBank. Data obtained for the 4 molecular markers evaluated were compared exclusively with congeneric species. Alignment constructions were performed using the algorithm MUSCLE implemented in the software MEGA X (Kumar et al., ). Before phylogenetic analyses, alignments were trimmed to match the shortest sequence. The evolutionary models used for the analyses were estimated by the Bayesian information criterion implemented in MEGA X. Outgroups for each dataset were chosen based on previous molecular phylogenetic studies including representatives of Echinostoma and related taxa (Sereno-Uribe et al., ; Tkach et al., ; Georgieva et al., ; Nakao and Sasaki, ). The methods used to generate the phylogenetic hypotheses were maximum likelihood (ML) and Bayesian inference (IB). ML analyses were performed in MEGA X and nodal support was measured using the bootstrap test with 1000 replicates. BI analyses were performed with MrBayes v.3.2.6 software (Ronquist et al., ) and posterior probability values were determined by the Monte Carlo method via Markov chains, in 2 simultaneous runs of 4 chains for 106 generations, with topologies sampling every 100 generations. The ‘burn-in’ was established for the first 25% of the trees sampled, and the nodal support was estimated from the remaining trees. The trees obtained by BI were visualized with FigTree software version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and edited in Adobe Photoshop® v.22.4.3 software (Adobe Inc., USA). All sequences generated in this study were deposited in GenBank under the accession numbers OQ132569 (28S), OQ132538 (ITS1-5.8S-ITS2), OQ134393 (cox1) and OQ126877 (nad1).
Results
A total of 56 specimens of a 37-collar-spined Echinostoma were recovered from the small intestine of M. unguiculatus 15 days after the experimental infection with metacercariae, obtained from S. marmorata. The obtained morphological and molecular data pointed to a new species of Echinostoma from the ‘revolutum’ group, which is described below.
Family Echinostomatidae Looss, 1899
Genus Echinostoma Rudolphi, 1809
Echinostoma maldonadoi Valadão, Alves & Pinto n. sp.
Natural intermediate host: Stenophysa marmorata (Guilding, 1828) (Gastropoda: Physidae)
Prevalence of infection: 1/224 (0.45%).
Experimental definitive host: Meriones unguiculatus (Milne-Edwards, 1867) (Rodentia: Muridae).
Natural definitive host: unknown.
Type locality: Alvorada de Minas (18°43′7″ S 43°22′5″ W), Minas Gerais, Brazil
Type material: Holotype (CHIOC 39915 a); paratypes (5 specimens, CHIOC 39915 b-f and 10 specimens, UFMG–TRE UFMG-TRE 133).
Representative DNA sequences: 28S (OQ132569), ITS1-5.8S-ITS2 (OQ132538), cox1 (OQ134393) and nad1 (OQ126877).
ZooBank registration: urn:lsid:zoobank.org:pub:2A535C25-8E19-4716-8E6F-B94AC683A7C6
Etymology: The specific name maldonadoi is given to honour the Brazilian helminthologist, Dr. Arnaldo Maldonado Júnior, for his relevant contributions to the biology and taxonomy of Echinostoma.
Description
Adult (n = 24, Figs 1 and 2; measurements in Table 1): Body slender, elongate, dorsoventrally flattened, with maximum width at uterine level, 4.5–6.4 times longer than wide, with almost parallel lateral margins and notable constriction at posterior margin of ventral sucker. Tegument with scale-like spines in alternating transverse rows, denser at pharynx region, extending to level of posterior margin of ventral sucker and reaching posterior testis level on ventral surface. Forebody short, 9–15% of body length. Head collar (n = 10) well developed, reniform, width representing 18–34% of maximum body width. Collar spines conspicuous, 37 in number in most specimens (5 with 37, 2 with 33, 1 with 36, 38 or 39 spines), with following arrangement: 5 angle spines (3 oral and 2 aboral); 6 lateral spines in single row on each ventral lappet, and 15 dorsal spines in double row (8 oral and 7 aboral). Dorsal spines varying in length, with the dorsolateral spines markedly larger than central (dorsalmost) spines. Oral sucker ventro-subterminal, rounded, muscular. Prepharynx very short; pharynx oval to elongate-oval, muscular. Oesophagus short, 2–4% of body length; oesophageal bifurcation just anterior to ventral sucker; intestinal caeca blind, almost reaching posterior extremity. Ventral sucker spherical, muscular, located in the first quarter of body; 2 times as large as oral sucker (suckers–width ratio 1:2.1–2.4). Testes 2, subglobular, smooth or slightly indented, in tandem, located in third quarter of body; post-testicular region (between posterior testis and posterior extremity) corresponding to 25–44% of body length. Cirrus sac elongate-oval, anterior to ventral sucker, with a simple elongate-oval seminal vesicle, well-developed pars prostatica, coiled ejaculatory duct and unspined cirrus. Genital pore at the level of anterior margin of ventral sucker. Ovary transversely oval, post-equatorial, pretesticular. Mehlis' gland contiguous with ovary. Uterus intercaecal, with numerous coils between ventral sucker and ovary; uterine field corresponding to 24–32% of body length. Metraterm short, weakly muscular. Vitellarium follicular, forming 2 lateral non-confluent fields of small follicles overlapping caeca and extending from level of the region slightly posterior to ventral sucker to posterior extremity of body. Eggs abundant, oval, yellow, operculated, immature.
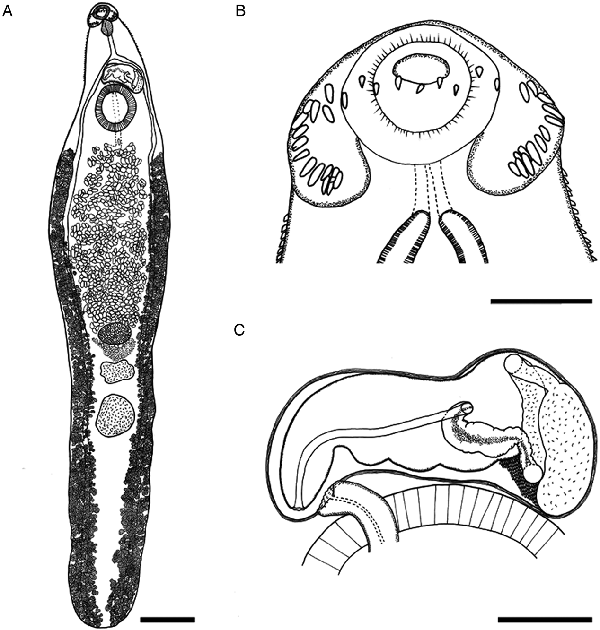
Fig. 1
Echinostoma maldonadoi n. sp. line drawings of the holotype. (A) Whole view. (B) Detail of cephalic collar. (C) Detail of genital complex. Scale bars: A: 1 mm; B, C: 250 μm.
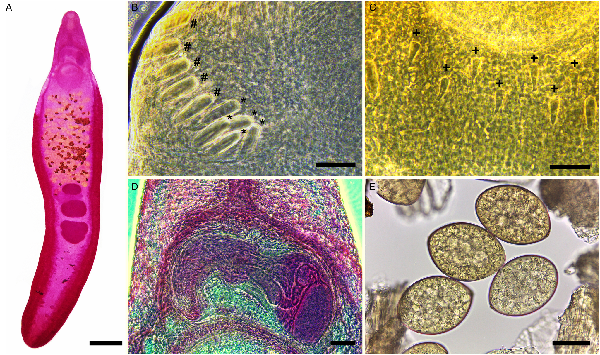
Fig. 2
Micrographs of Echinostoma maldonadoi n. sp. (A) Whole view of a carmine-stained paratype. (B) Detail of angular (*) and lateral (#) spines. (C) Detail of dorsal spines (+). (D) Detail of the cirrus sac. (E) Eggs obtained in feces. Scale bars: A: 1 mm; B, C, E: 50 μm; D: 100 μm.
Larval stages
Mature redia (n = 23, Fig. 3A; Table 2): Body elongate, tapered at its anterior and posterior ends, with yellowish pigment. Collar absent. Pharynx subspherical, muscular. Caecum short, with dark pigment inside. Birth pore prominent at the anterior region, lateral. Two prominent locomotor appendages present at about 2/3 of body length.
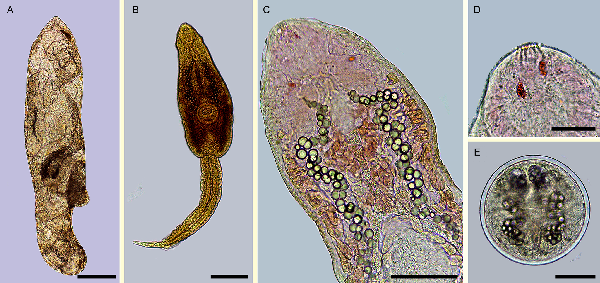
Fig. 3
Echinostoma maldonadoi n. sp. larval stages found in Stenophysa marmorata from Brazil. (A) Redia. (B) Cercaria. (C) Detail of the body of cercaria. (D) Detail of anterior end. (E) Metacercaria. Scale bars: A: 200 μm; B: 100 μm; C, E: 50 μm; D: 20 μm.
Cercaria (n = 36, Fig. 3B–D; Table 2): Body elongate-oval, with maximum width just anterior to ventral sucker. Tegument smooth. Collar well developed, with 37 spines, arranged as in adults. Oral sucker rounded, subterminal. Pharynx ovoid, muscular. Oesophagus long; oesophageal bifurcation just anterior to ventral sucker. Caeca long, with granular content inside, extending up to posterior extremity. Ventral sucker subglobular, muscular, just post-equatorial, slightly larger than the oral sucker; average ratio between suckers width, 1:1.4 (1:1.0–1.6). Penetration glands with granular content, preacetabular, medially distributed, along oesophagus, with canaliculi directed to anterior region, opening in 6 ducts at extremity of oral sucker (Fig. 3D). Paraesophagean glands absent. Cystogeneous glands with rhabditiform content located laterally through body. Genital primordia composed by 2 masses of cells dorsal to ventral sucker. Tail long, larger than body, average ratio between body and tail length 1:1.4 (1:0.9–1.7); ending in a highly contractile finger-like process, slender, presenting tegumental fin-folds. Number and disposition of tail fin-folds not observed. Excretory vesicle small, at posterior extremity of body. Main ducts of excretory system presenting numerous, small calcareous concretions (about 50, 6–9 μm of diameter) between the pharynx and ventral sucker. Details of excretory system, including flame-cell formula, position of excretory pore and caudal excretory duct not characterized.
Metacercaria (n = 35, Fig. 3E; Table 2): cysts spherical, on average 135 ± 7 μm in diameter; cyst walls composed of 2 layers, the outer layer transparent, about 6 μm thick, and the inner layer opaque, thin, about 2 μm thick.
Remarks
The experimentally recovered adult specimens can be readily distinguished from all but 1 species of Echinostoma of the ‘revolutum’ complex by bearing tiny dorsal collar spines, much smaller than the dorsolateral and lateral ones (see description). Despite the differences in the head collar commented above, the recently described E. pseudorobustum Valadão et al., 2022 presents similar size and body shape. However, this species is differentiated by having a longer forebody (FORE% c. 18.9 vs 12), oesophagus length as a proportion of body length (OSL% c. 10 vs 3), head collar as a proportion of body width (CW/BW c. 49.5 vs 26.1%) and uterine field (OVAR c. 41.6 vs 28%).
Taking into account the morphological data alone, we initially assigned the new taxon to E. paraensei because this species is the only one in the ‘revolutum’ complex that also possesses tiny dorsal collar spines (Kostadinova and Gibson, ), and also due to similar morphometrics comparing other key morphological features (see Table 1). The geographical origin of the specimens studied (Mesoregion of Belo Horizonte) also led us to preliminarily identify the isolate as E. paraensei. There are some differences observed between the newly collected specimens and those previously identified as E. paraensei by other workers, e.g. body shape (BW%, 19 vs 12–13), ratio between the width of oral and ventral suckers (OSW:VSW, 1:2.5 vs 1:1.4–2.8) and prepharynx length (PL) (PL: 41 vs 72–120 μm in the original description). These differences are attributed here to the evaluation of specimens with different ages (range of 12–109 dpi) in each study as individuals continue to grow after reaching maturity and the use of different techniques for fixing the material (flattening or not of the specimens).
While morphological features suggested the conspecificity of E. maldonadoi n. sp. and E. paraensei, molecular data, specifically based on the mitochondrial nad1 gene, revealed interspecific levels of nucleotide divergence between the 2 species (see below).
Molecular study
Partial sequences of the 28S rRNA gene (1197 bp), ITS1-5.8S-ITS-2 region (1159 bp), cox1 gene (415 bp) and nad1 gene (478 bp) were successfully generated for the isolate of 37-collar-spined E. maldonadoi n. sp. Phylogenetic trees were reconstructed using BI and ML criteria, but only those obtained through the first approach are shown, including values of nodal support for both analyses. The trimmed 28S alignment was 1051 bp long and included 18 sequences for 14 species of the ‘revolutum’ group of Echinostoma. The phylogenetic reconstruction based on this gene (Fig. 4A) revealed that E. maldonadoi n. sp. clustered in a well-supported clade with an isolate of E. paraensei maintained under laboratory conditions in the USA, without nucleotide differences between the sequences. The interspecific variation in relation to other Echinostoma spp. included in the analyses was 0.3–1.1%.
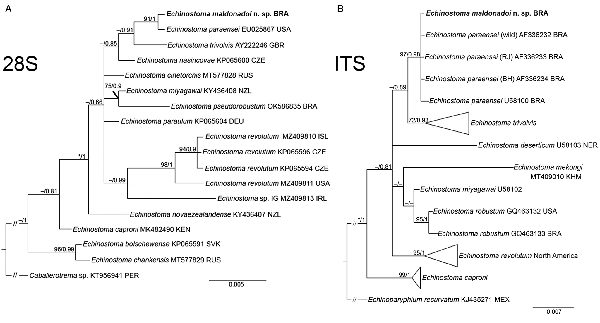
Fig. 4
Phylogenetic relationship between Echinostoma maldonadoi n. sp. (in bold) and other 37-collar-spined Echinostoma species inferred from sequences of 28S rDNA (1051 bp; nucleotide substitution model: HKY + G) (A) and of internal transcribed spacer (ITS) (1001 bp; nucleotide substitution model: K2 + G) (B) based on maximum likelihood (ML) and Bayesian inference (BI) analyses. Nodal support is indicated as ML/BI; values <0.70 (BI) and <70 (ML) are indicated by a dash.
A dataset containing 9 species of Echinostoma was included in the analysis of the nuclear region ITS1-5.8S-ITS2 (trimmed alignment: 1001 bp). Echinostoma maldonadoi n. sp. grouped with 3 Brazilian isolates of E. paraensei in a strongly supported clade (97/0.98) (Fig. 4B), but the actual phylogenetic affinities with these isolates remained unclear. Moreover, the molecular divergence in relation to the 4 isolates of E. paraensei from N. squamipes was low (0–0.1%). Representatives of this clade appeared as sister to isolates of E. trivolvis, but the support was low. The interspecific genetic variation in relation to other Echinostoma spp. included in the analyses was 0.7–2.6%.
The molecular data obtained for the faster evolving mitochondrial genes (nad1 and cox1) revealed nucleotide differences between the new species and E. paraensei. For cox1 gene, a trimmed alignment (366 bp) containing 8 species of Echinostoma was analysed. The resolution of the tree was poor resulting in a polytomy, which hampered our understanding on the phylogenetic relationships of most taxa (Fig. 5A). Nevertheless, E. maldonadoi n. sp. was recovered in a strongly supported clade together with E. paraensei. Moreover, in the cox1 tree, E. maldonadoi n. sp. + E. paraensei is a sister group to E. caproni, and this nesting was well-supported by BI (0.96). This clade forms a polytomy with E. cinetorchis, E. miyagawai and 2 ‘E. revolutum’ isolates. The genetic divergence between E. maldonadoi n. sp. and the laboratory isolate of E. paraensei from the USA was 3.8% (difference at 14 nucleotide positions), which might suggest that they do not belong to the same species. The molecular divergence in relation to other Echinostoma spp. ranged from 10.9 to 15.9%.
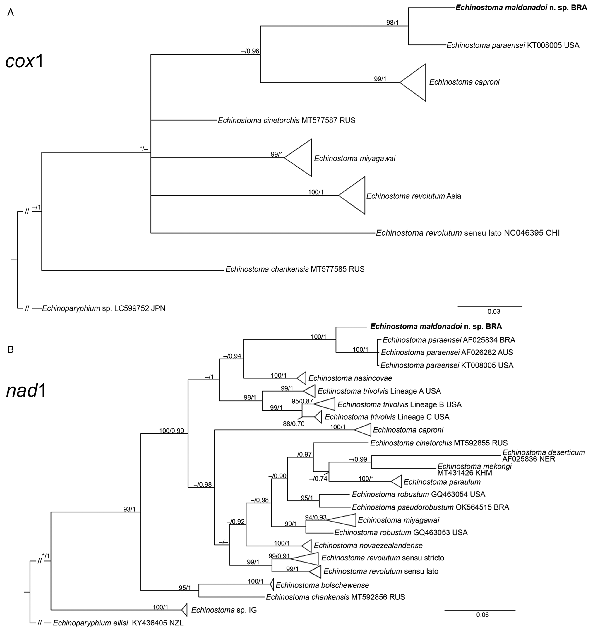
Fig. 5
Phylogenetic relationship between Echinostoma maldonadoi n. sp. (in bold) and other 37-collar-spined Echinostoma species inferred from sequences of cox1 mtDNA (366 bp; nucleotide substitution model: HKY + I) (A) and of nad1 (438 bp; nucleotide substitution model: HKY + G + I) (B) based on maximum likelihood (ML) and Bayesian inference (BI) analyses. Nodal support is indicated as ML/BI; values <0.70 (BI) and <70 (ML) are indicated by a dash.
The molecular analysis based on the nad1 gene unequivocally confirmed the distinct species status of E. maldonadoi n. sp. The trimmed alignment (438 bp) included 153 sequences of 19 species/species-level lineages of Echinostoma. In the well-resolved phylogenetic tree (Fig. 5B), the new species grouped in a strongly supported clade with 3 isolates of E. paraensei, and both appeared as sister clade to E. nasincovae (albeit with no support in the ML analysis). The genetic divergence in relation to the isolate of E. paraensei maintained under laboratory conditions in the USA, and in relation to another isolate of the same species from Brazil, reported by Morgan and Blair (), was 6.2% (for 2 isolates) and 6.4% (27 and 28 nucleotides), respectively, which fell into the interspecific range of variability for Echinostoma spp. of the ‘revolutum’ group. Moreover, the newly described species exhibited a genetic divergence of 12.3–21.0% compared with the other species of the ‘revolutum’ group. Echinostoma maldonadoi n. sp. can be diagnosed based on unique sites/nucleotides in specific positions of the nad1 alignment comparing to isolates of E. paraensei: 11–A, 20–C, 36–A, 37–T, 47–A, 68–G, 71–C, 81–T, 107–C, 119–A, 122–T, 144–A, 188–G, 197–T, 245–A, 260–T, 275–A, 314–T, 321–A, 337–C, 351–C, 365–T, 372–G, 398–G, 402–C, 405–G, 410–T (see Supplementary file – Alignment of sequences nad1 from these isolates).
Discussion
Our understanding of the species diversity of the genus Echinostoma has been improved by integrating molecular and morphological approaches to better delimit species boundaries and consequently address more accurately questions concerning host use, geographical distribution and life cycle (Georgieva et al., , ; Faltýnková et al., ; Izrailskaia et al., ; Valadão et al., ). A number of studies stressed the presence of several cryptic lineages within the ‘revolutum’ species complex of Echinostoma, mostly from the Holarctic realm (Detwiler et al., , ; Georgieva et al., ; Izrailskaia et al., ), which presented challenges either for identification of known taxa or the proposal of new taxa with no morphological differential trait to report. Even though cryptic diversity is more common among trematodes than other groups of parasitic helminths (Pérez-Ponce de León and Poulin, ), some of the cryptic evolutionary independent lineages found in previous studies of Echinostoma spp. may be found morphologically identifiable, once all developmental stages (primarily adults) are described. In the present study, the integration of morphological, molecular and life cycle data made it possible to propose a new species of Echinostoma, E. maldonadoi n. sp., cryptically related to E. paraensei considering all developmental stages, yet some morphometric differences were found between isolates of the new species and E. paraensei (see remarks).
From the biological point of view, the intermediate host use is not considered a significant difference between E. paraensei and E. maldonadoi n. sp. Although natural infection for the former was reported in planorbid snails, B. grabrata and Glyptophysa sp., in Brazil and Australia, respectively (Lie and Basch, ; Morgan and Blair, ), the experimental susceptibility of physid snails to E. paraensei was reported to the isolate from Belo Horizonte (original description) (Lie and Basch, ) and for the isolate from Sumidouro, state of Rio de Janeiro, Brazil (Maldonado et al., ). Nevertheless, echinostome cercariae were not found among 767 specimens of Biomphalaria spp. (including B. glabrata) collected in Alvorada de Minas (Coelho et al., ), the type locality of E. maldonadoi n. sp., which may suggest that E. paraensei and E. maldonadoi n. sp. are primarily found in planorbid and physid snails, respectively, at least in natural conditions.
A question that emerges from this study, but that is impossible to be answered at the current stage of the knowledge, is whether the subsequent reports of E. paraensei-like specimens really correspond to this taxon or E. maldonadoi n. sp., or even to another new cryptic species of the genus. Interestingly, some relevant differences between isolates identified as E. paraensei were reported regarding: the time of miracidial development, worm burden, number of uterine eggs and parasite distribution in the site of infection (Maldonado et al., , , ). The possibility that such differences may represent interspecific variation instead of just evident phenotypic plasticity, as previously thought (Maldonado et al., ), cannot be ruled out. In this sense, future nad1 sequencing of this laboratory-maintained isolate of E. paraensei is here encouraged for taxonomic validation of the parasite species used in the different experimental studies previously published (Ferraz et al., ; Gonçalves et al., ; Monte et al., , , ; Souza et al., ).
The utility of the most densely sampled nuclear ribosomal genetic regions, 28S and ITS, and mitochondrial protein-coding genes, cox1 and nad1, were evaluated in molecular phylogenetic studies (Morgan and Blair, , ; Georgieva et al., ; Izrailskaia et al., ). While all of them agree that mitochondrial genes should be the first choice for barcoding species of Echinostoma in the ‘revolutum’ group, as nuclear ribosomal genes may not provide enough phylogenetic signal to differentiate closely related species, they differ concerning which mitochondrial marker is more informative. Most of the scientific contributions provided compelling evidence that nad1 shows a sufficient gap between levels of intraspecific and interspecific genetic divergence, the so-called barcode gap, allowing the identification of species boundaries (Morgan and Blair, ; Georgieva et al., , ). For this reason, most taxonomic surveys on Echinostoma provided nad1 sequences (Georgieva et al., , ; Faltýnková et al., ; Pantoja et al., ). However, Izrailskaia et al. () argued that cox1 may be more informative than nad1 and might solve more confidently taxonomic problems within the Echinostomatidae.
In the present study, we reinforce the limitations of ribosomal nuclear DNA for the distinction of closely related species (the new taxon could not be recognized using this dataset) and found only moderate nucleotide divergence (3.83%) using cox1 between the sister taxa E. maldonadoi n. sp. and E. paraensei, much lower than the 6.3–13.7% and 7.3–16.8% interspecific values reported by Morgan and Blair () and Izrailskaia et al. (), respectively. These different values may just be biased by the paucity of cox1 sequences for Echinostoma spp., leaving just a few sequences of closely related sister taxa to be compared. Contrary to that, the divergence values (6.2–6.4%) of the nad1 marker between the new species and E. paraensei were within the interspecific range of variability for Echinostoma spp. of the ‘revolutum’ group. For instance, in the most comprehensive study involving molecular taxonomy and species delineation in Echinostoma spp., Georgieva et al. () recognized 17 species/species-level lineages using nad1 dataset and found 4.2–21.5% of interspecific divergence. Moreover, identification was achieved for all monophyletic lineages at 3% divergence threshold. These same authors found a mean intraspecific divergence of 0.2–1.8%. More recently, Pantoja et al. () reported 0–1.6% of intraspecific divergence for Echinostoma revolutum s.str. from Europe and 0.5–1.2% for isolates identified as Echinostoma sp. IG. Thus, the divergence found between E. maldonadoi n. sp. and E. paraensei is more than double from the higher intraspecific divergence previously reported for species of Echinostoma. It is also worth noting, that the nucleotide divergence using nad1 between E. maldonadoi n. sp. and the remaining species of the ‘revolutum’ group (12.3–21.0%) was considerably higher than compared with its sister taxon (E. paraensei), which may indicate a very recent speciation event and segregation into these 2 morphologically indistinguishable species.
Our findings support the conclusions of Morgan and Blair () and Georgieva et al. () that the best mitochondrial marker for the investigation of phylogenetic interrelationships and identification of cryptic diversity within the ‘revolutum’ group of Echinostoma is nad1. In fact, most internal or terminal clades are strongly supported in our phylogenetic reconstruction. Moreover, the availability of nad1 datasets with larger taxonomic and geographical coverage in relation to the remaining markers allows the comparison of genetic distances between 2 putative species with values available for ‘benchmark’ congeners. Nevertheless, finding a ‘genetic yardstick’ remains challenging in trematodes (Nadler and Pérez-Ponce de León, ) and even in Echinostoma.
Even though the reciprocal monophyly of E. maldonadoi n. sp. and E. paraensei could not be assessed as only a single haplotype of the new taxon was herein detected, the knowledge leveraged for the molecular taxonomy of this genus in the last 2 decades, including a clear barcode gap using nad1 dataset, suffices for our new species proposal. This hypothesis though may be strengthened by molecular characterization of new isolates and widening the number of target molecular markers, such as encompassing the entire mitochondrial genome.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S003118202300001X
References
- Adema CM, Léonard PM, DeJong RJ, Day HL, Edwards DJ, Burgett G, Hertel LA and Loker ES (2000) Analysis of messages expressed by Echinostoma paraensei miracidia and sporocysts, obtained by random EST sequencing. Journal of Parasitology 86, 60–65.
- Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215, 403–410.
- Chai JY, Cho J, Chang T, Jung BK and Sohn WM (2020) Taxonomy of Echinostoma revolutum and 37-collar-spined Echinostoma spp.: a historical review. Korean Journal of Parasitology 58, 343–371.
- Chai JY, Jung BK, Chang T, Shin H, Cho J, Ryu JY, Kim HS, Park K, Jeong MH, Hoang EH and Abullah MBM (2021) Echinostoma miyagawai Ishii, 1932 (Echinostomatidae) from ducks in Aceh Province, Indonesia with special reference to its synonym with Echinostoma robustum Yamaguti, 1935. Korean Journal of Parasitology 59, 35–45.
- Coelho PRS, Ker FTO, Araújo AD, Guimarães RJPS, Negrão-Corrêa DA, Caldeira RL and Geiger SM (2021) Identification of risk areas for intestinal schistosomiasis, based on malacological and environmental data and on reported human cases. Frontiers in Medicine 8, 642348.
- Coelho PRS, Ker FTO, Araújo AD, Pinto HA, Negrão-Corrêa DA, Caldeira RL and Geiger SM (2022) Ecological aspects and survey of limnic gastropods: relationships between human health and conservation. Pathogens 11, 1533.
- Combes C (1982) Trematodes: antagonism between species and sterilizing effects on snails in biological control. Parasitology 84, 151–175.
- Detwiler JT, Bos DH and Minchella DJ (2010) Revealing the secret lives of cryptic species: examining the phylogenetic relationships of echinostome parasites in North America. Molecular Phylogenetics and Evolution 55, 611–620.
- Detwiler JT, Zajac AM, Minchella DJ and Belden LK (2012) Revealing cryptic parasite diversity in a definitive host: echinostomes in muskrats. Journal of Parasitology 98, 1148–1155.
- Faltýnková A, Georgieva S, Soldánová M and Kostadinova A (2015) A re-assessment of species diversity within the ‘revolutum’ group of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) in Europe. Systematic Parasitology 90, 1–25.
- Fernandes BMM, Justo MCN, Cárdenas MQ and Cohen SC (2015) South American Trematodes Parasites of Birds and Mammals. Rio de Janeiro: Oficina de Livros.
- Ferraz J, Souza J, Costa-Silva M, Torres E, Santana A, Lanfredi R, Maldonado Jr A and Garcia J (2012) Effect of praziquantel on adult Echinostoma paraensei worms in experimentally infected mice. Parasitology Research 111, 143–148.
- Fried B and Graczyk TK (2000) Echinostomes as Experimental Models for Biological Research. Dordrecht: Kluwer Academic Publishers.
- Fried B and Peoples RC (2009) Maintenance, cultivation, and excystation of echinostomes: 2000–2007. In Fried B and Toledo R (eds), The Biology of Echinostomes: From the Molecule to the Community. New York: Springer Science + Business Media, pp. 111–128.
- Fried B, Kanev I and Reddy A (2009) Studies on collar spines of echinostomatid trematodes. Parasitology Research 105, 605–608.
- Fujino T, Nakano T, Washioka H, Tonosaki A, Ichikawa H and Fried B (2000) Comparative ultrastructure of eggs in Echinostoma paraensei, E. caproni and E. trivolvis (Trematoda: Echinostomatidae). Parasitology Research 86, 427–430.
- Garcia JS, Maldonado Jr A, Bidau CJ, Corrêa LR, Lanfredi RM and Coelho PMZ (2010) The effect of early infection with Echinostoma paraensei on the interaction of Schistosoma mansoni with Biomphalaria glabrata and Biomphalaria tenagophila. Memórias do Instituto Oswaldo Cruz 105, 499–503.
- Garcia JS, Hooper CS, Simões RO, Santos MAJ, Maldonado Jr A and Silva JP (2011). Biochemical and histological responses of Rattus norvegicus (Wistar) infected with Echinostoma paraensei (Trematoda: Echinostomatidae). Veterinary Parasitology 178, 86–92.
- Garcia JS, Simões RO, Mota E, Santos IS and Maldonado Jr A (2021) Pathological effects of Echinostoma paraensei infection associated with pyogenic abscesses caused by Pseudomonas aeruginosa in Swiss mice. Revista de Patologia Tropical 50, 40–48.
- Georgieva S, Selbach C, Faltýnková A, Soldánová M, Sures B, Skirnisson K and Kostadinova A (2013) New cryptic species of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasites & Vectors 6, 64.
- Georgieva S, Faltýnková A, Brown R, Blasco-Costa I, Soldánová M, Sitko J, Scholz T and Kostadinova A (2014) Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasites & Vectors 7, 520.
- Georgieva S, Blasco-Costa I and Kostadinova A (2017) Molecular characterization of four echinostomes (Digenea: Echinostomatidae) from birds in New Zealand, with descriptions of Echinostoma novaezealandense n. sp. and Echinoparyphium poulini n. sp. Systematic Parasitology 94, 477–497.
- Gonçalves JP, Oliveira-Menezes A, Maldonado Jr A, Carvalho TM and Souza W (2013) Evaluation of praziquantel effects on Echinostoma paraensei ultrastructure. Veterinary Parasitology 194, 16–25.
- Hanington PC, Lun CM, Adema CM and Loker ES (2010) Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. International Journal for Parasitology 40, 819–831.
- Izrailskaia AV, Besprozvannykh VV and Tatonova YV (2021) Echinostoma chankensis nom. nov., other Echinostoma spp. and Isthmiophora hortensis in East Asia: morphology, molecular data and phylogeny within Echinostomatidae. Parasitology 148, 1366–1382.
- Kostadinova A (2005) Family Echinostomatidae Looss, 1899. In Jones A, Gibson DI and Bray R (eds), Keys to the Trematoda, vol. 2. London, UK: CABI Publishing and The Natural History Museum, pp. 9–64.
- Kostadinova A and Gibson DI (2000) The systematics of the echinostomes. In Fried B and Graczyk TK (eds), Echinostomes as Experimental Models for Biological Research. Dordrecht: Kluwer, pp. 31–57.
- Kostadinova A, Herniou EA, Barret J and Littlewood DTJ (2003) Phylogenetic relationships of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) and related genera re-assessed via DNA and morphological analyses. Systematic Parasitology 54, 159–176.
- Kumar S, Stecher G, Li M, Knyaz C and Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549.
- Lie KJ and Basch PF (1966) Life history of Echinostoma barbosai sp. n. (Trematoda: Echinostomatidae). Journal of Parasitology 52, 1052–1057.
- Lie KJ and Basch PF (1967) The life history of Echinostoma paraensei sp. n. (Trematoda: Echinostomatidae). Journal of Parasitology 53, 1192–1199.
- Loker ES, Cimino DF, Stryker GA and Hertel LA (1987) The effect of size of M-line Biomphalaria glabrata on the course of development of Echinostoma paraensei. Journal of Parasitology 73, 1090–1098.
- Loker ES, Cimino DF and Hertel LA (1992) Excretory-secretory products of Echinostoma paraensei mediate interference with Biomphalaria glabrata hemocyte functions. Journal of Parasitology 78, 104–115.
- Lunaschi LI, Drago FB and Nunez V (2018) Two new species of Echinostoma (Digenea: Echinostomatidae) from Argentinean birds. Revista Mexicana de Biodiversidade 89, 356–364.
- Luton K, Walker D and Blair D (1992) Comparison of ribosomal internal transcribed spacer from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea). Molecular and Biochemical Parasitology 56, 323–328.
- Lutz A (1919) O Schistosomum mansoni e a schistosomatose, segundo observações feitas no Brasil. Memórias do Instituto Oswaldo Cruz 11, 121–155.
- Maldonado Jr A, Loker ES, Morgan JAT, Rey L and Lanfredi RM (2001a) Description of the adult worms of a new Brazilian isolate of Echinostoma paraensei (Platyhelminthes: Digenea) from its natural vertebrate host Nectomys squamipes by light and scanning electron microscopy and molecular analysis. Parasitology Research 87, 840–848.
- Maldonado Jr A, Vieira GO, Garcia JS, Rey L and Lanfredi RM (2001b) Biological aspects of a new isolate of Echinostoma paraensei (Trematoda: Echinostomatidae): susceptibility of sympatric snails and the natural vertebrate host. Parasitology Research 87, 853–859.
- Maldonado Jr A, Vieira GO and Lanfredi RM (2003) Echinostoma luisreyi n. sp. (Platyhelminthes: Digenea) by light and scanning electron microscopy. Journal of Parasitology 89, 800–808.
- Maldonado Jr A, Zeitone BK, Amado LA, Rosa IF, Machado-Silva JR and Lanfredi RM (2005) Biological variation between two Brazilian geographical isolates of Echinostoma paraensei. Journal of Helminthology 79, 345–351.
- Marcilla A (2009) Echinostomes: genomics and proteomics. In Fried B and Toledo R (eds), The Biology of Echinostomes: From the Molecule to the Community. New York: Springer Science + Business Media, pp. 130–141.
- Miura O, Kuris AM, Torchin ME, Hechinger RF, Dunham EJ and Chiba S (2005) Molecular-genetic analyses reveal cryptic species of trematodes in the intertidal gastropod, Batillaria cumingi (Crosse). International Journal for Parasitology 35, 793–780.
- Monte TCC, Garcia JS, Gentile R, Vasconcellos MC, Souza J, Braga BV and Maldonado Jr A (2016) In vivo and in vitro effects of the herbicide Roundup (®) on developmental stages of the trematode Echinostoma paraensei. Experimental Parasitology 169, 43–50.
- Monte TCC, Braga BV, Vasconcellos MC, Jurberg AD, Mota EM, Barbosa HS, Garcia JS and Maldonado Jr A (2018) Morphological effects on helminth parasites caused by herbicide under experimental conditions. Revista Brasileira de Parasitologia Veterinária 27, 42–51.
- Monte TCC, Chometon TQ, Bertho AL, Moura VS, Vasconcellos MC, Garcia J, Ferraz-Nogueira R, Maldonado Jr A and Faro MJ (2019) Changes in hemocytes of Biomphalaria glabrata infected with Echinostoma paraensei and exposed to glyphosate-based herbicide, Journal of Invertebrate Pathology 160, 67–75.
- Morgan JAT and Blair D (1998a) Mitochondrial ND1 gene sequences used to identify echinostome isolates from Australia and New Zealand. International Journal for Parasitology 28, 493–502.
- Morgan JAT and Blair D (1998b) Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology 116, 289–297.
- Nadler S and Pérez-Ponce de León G (2011) Integrating molecular and morphological approaches for characterizing parasite cryptic species: implications for parasitology. Parasitology 138, 1688–1709.
- Nakao M and Sasaki M (2021) Trematode diversity in freshwater snails from a stopover point for migratory waterfowls in Hokkaido, Japan: an assessment by molecular phylogenetic and population genetic analyses. Parasitology International 83, 102329.
- Nollen PM (1996) The mating behavior of Echinostoma paraensei grown in mice. Journal of Helminthology 70, 43–45.
- Nowak TS and Loker ES (2005) Echinostoma paraensei: differential gene transcription in the sporocyst stage. Experimental Parasitology 109, 94–105.
- Pantoja C, Faltýnková A, O'Dwyer K, Jouet D, Skírnissin K and Kudlai O (2021) Diversity of echinostomes (Digenea: Echinostomatidae) in their snail hosts at high latitudes. Parasite 28, 59.
- Pérez-Ponce de León G and Poulin R (2018) An updated look at the uneven distribution of cryptic diversity among parasitic helminths. Journal of Helminthology 92, 197–202.
- Pinheiro J, Maldonado Jr A, Attias M and Lanfredi RM (2005) Ultrastructure of the miracidium of Echinostoma paraensei Lie and Basch, 1967 (Trematoda, Echinostomatidae). Parasitology Research 97, 367–372.
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA and Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542.
- Sereno-Uribe AL, Pinacho-Pinacho CD, Sanchéz Cordero V and García-Varela M (2015) Morphological and molecular analyses of larval and adult stages of Echinoparyphium recurvatum von Linstow 1873 (Digenea: Echinostomatidae) from central Mexico. Journal of Helminthology 89, 458–464.
- Souza JGR, Garcia JS, Gomes APN, Machado-Silva JR and Maldonado Jr A (2017a) Comparative pattern of growth and development of Echinostoma paraensei (Digenea: Echinostomatidae) in hamster and Wistar rat using light and confocal laser scanning microscopy. Experimental Parasitology 13, 41–49.
- Souza JGR, Lopes-Torres EJ, Garcia JS, Gomes APN, Rodrigues-Silva R, Maldonado Jr A and Machado-Silva JR (2017b) Light and scanning electron microscopy study of in vitro effects of artesunate in newly excysted metacercariae of Echinostoma paraensei (Trematoda: Digenea). Experimental Parasitology 174, 10–16.
- Sullivan JT and Richards CS (1981) Schistosoma mansoni, NIH-SM-PR-2 strain, in susceptible and nonsusceptible stocks of Biomphalaria glabrata: comparative histology. Journal of Parasitology 67, 702–708.
- Tkach VV, Littlewood DTJ, Olson PD, Kinsella JM and Swiderski Z (2003) Molecular phylogenetic analysis of the Microphalloidea Ward, 1901 (Trematoda: Digenea). Systematic Parasitology 56, 1–15.
- Tkach VV, Kudlai O and Kostadinova A (2016) Molecular phylogeny and systematics of the Echinostomatoidea Looss, 1899 (Platyhelminthes: Digenea). International Journal for Parasitology 46, 1–15.
- Toledo R (2009) Echinostomes in the definitive host: a model for the study of host-parasite relationships. In Fried B and Toledo R (eds), The Biology of Echinostomes: From the Molecule to the Community. New York: Springer Science + Business Media, pp. 89–110.
- Toledo R and Fried B (2005) Echinostomes as experimental models for interactions between adult parasites and vertebrate hosts. Trends in Parasitology 21, 251–254.
- Toledo R, Esteban JG and Fried B (2009) Recent advances in the biology of echinostomes. Advances in Parasitology 69, 147–204.
- Travassos L, Teixeira de Freitas JF and Kohn A (1969) Trematódeos do Brasil. Memórias do Instituto Oswaldo Cruz 67, 1–886.
- Valadão MC, López-Hernández D, Alves PV and Pinto HA (2022) A new species of Echinostoma (Trematoda: Echinostomatidae) from the ‘revolutum’ group found in Brazil: refuting the occurrence of Echinostoma miyagawai (=E. robustum) in the Americas. Parasitology 149, 325–336.